Middle East respiratory syndrome coronavirus vaccine based on a propagation-defective RNA replicon elicited sterilizing immunity in mice
- PMID: 34686605
- PMCID: PMC8639359
- DOI: 10.1073/pnas.2111075118
Middle East respiratory syndrome coronavirus vaccine based on a propagation-defective RNA replicon elicited sterilizing immunity in mice
Abstract
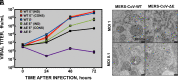
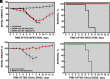
Self-amplifying RNA replicons are promising platforms for vaccine generation. Their defects in one or more essential functions for viral replication, particle assembly, or dissemination make them highly safe as vaccines. We previously showed that the deletion of the envelope (E) gene from the Middle East respiratory syndrome coronavirus (MERS-CoV) produces a replication-competent propagation-defective RNA replicon (MERS-CoV-ΔE). Evaluation of this replicon in mice expressing human dipeptidyl peptidase 4, the virus receptor, showed that the single deletion of the E gene generated an attenuated mutant. The combined deletion of the E gene with accessory open reading frames (ORFs) 3, 4a, 4b, and 5 resulted in a highly attenuated propagation-defective RNA replicon (MERS-CoV-Δ[3,4a,4b,5,E]). This RNA replicon induced sterilizing immunity in mice after challenge with a lethal dose of a virulent MERS-CoV, as no histopathological damage or infectious virus was detected in the lungs of challenged mice. The four mutants lacking the E gene were genetically stable, did not recombine with the E gene provided in trans during their passage in cell culture, and showed a propagation-defective phenotype in vivo. In addition, immunization with MERS-CoV-Δ[3,4a,4b,5,E] induced significant levels of neutralizing antibodies, indicating that MERS-CoV RNA replicons are highly safe and promising vaccine candidates.
Keywords: MERS-CoV; RNA replicon; coronavirus; vaccine.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: The authors declare patent applications (pending) filed by their institution.
Figures

Similar articles
-
Recombinant Receptor-Binding Domains of Multiple Middle East Respiratory Syndrome Coronaviruses (MERS-CoVs) Induce Cross-Neutralizing Antibodies against Divergent Human and Camel MERS-CoVs and Antibody Escape Mutants.J Virol. 2016 Dec 16;91(1):e01651-16. doi: 10.1128/JVI.01651-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795425 Free PMC article.
-
One-Health: a Safe, Efficient, Dual-Use Vaccine for Humans and Animals against Middle East Respiratory Syndrome Coronavirus and Rabies Virus.J Virol. 2017 Jan 3;91(2):e02040-16. doi: 10.1128/JVI.02040-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27807241 Free PMC article.
-
ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice.Vaccine. 2017 Jun 27;35(30):3780-3788. doi: 10.1016/j.vaccine.2017.05.032. Epub 2017 Jun 1. Vaccine. 2017. PMID: 28579232 Free PMC article.
-
Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism.Viruses. 2019 Jan 17;11(1):74. doi: 10.3390/v11010074. Viruses. 2019. PMID: 30658390 Free PMC article. Review.
-
Antibodies and vaccines against Middle East respiratory syndrome coronavirus.Emerg Microbes Infect. 2019;8(1):841-856. doi: 10.1080/22221751.2019.1624482. Emerg Microbes Infect. 2019. PMID: 31169078 Free PMC article. Review.
Cited by
-
Advances in Molecular Genetics Enabling Studies of Highly Pathogenic RNA Viruses.Viruses. 2022 Nov 30;14(12):2682. doi: 10.3390/v14122682. Viruses. 2022. PMID: 36560685 Free PMC article. Review.
-
The Reassessed Potential of SARS-CoV-2 Attenuation for COVID-19 Vaccine Development-A Systematic Review.Viruses. 2022 May 7;14(5):991. doi: 10.3390/v14050991. Viruses. 2022. PMID: 35632736 Free PMC article.
-
Alicante-Winter Immunology Symposium in Health (A-Wish) and the Boulle-SEI awards: A collaboration between the Spanish Society for immunology, the University of Alicante and the Jean Boulle Group to honor the Balmis Expedition.Curr Res Immunol. 2022 Jun 18;3:136-145. doi: 10.1016/j.crimmu.2022.06.001. eCollection 2022. Curr Res Immunol. 2022. PMID: 35754933 Free PMC article. Review.
-
Nature of viruses and pandemics: Coronaviruses.Curr Res Immunol. 2022;3:151-158. doi: 10.1016/j.crimmu.2022.08.003. Epub 2022 Aug 8. Curr Res Immunol. 2022. PMID: 35966177 Free PMC article. Review.
-
Construction and evaluation of a self-replicative RNA vaccine against SARS-CoV-2 using yellow fever virus replicon.PLoS One. 2022 Oct 20;17(10):e0274829. doi: 10.1371/journal.pone.0274829. eCollection 2022. PLoS One. 2022. PMID: 36264936 Free PMC article.
References
-
- International Committee on Taxonomy of Viruses, ICTV Master Species List 2019.v1 (2019). https://talk.ictvonline.org/files/master-species-lists/m/msl/9601. Accessed 9 June 2021.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

