The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation
- PMID: 34685640
- PMCID: PMC8534235
- DOI: 10.3390/cells10102660
The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation
Abstract
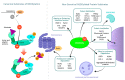
Neuronal precursor cell-expressed developmentally down-regulated protein 8 (NEDD8) is a ubiquitin-like protein (UBL) whose canonical function involves binding to, and thus, activating Cullin-Ring finger Ligases (CRLs), one of the largest family of ubiquitin ligases in the eukaryotic cell. However, in recent years, several non-canonical protein substrates of NEDD8 have been identified. Here we attempt to review the recent literature regarding non-canonical NEDDylation of substrates with a particular focus on how the covalent modification of NEDD8 alters the protein substrate. Like much in the study of ubiquitin and UBLs, there are no clear and all-encompassing explanations to satisfy the textbooks. In some instances, NEDD8 modification appears to alter the substrates localization, particularly during times of stress. NEDDylation may also have conflicting impacts upon a protein's stability: some reports indicate NEDDylation may protect against degradation whereas others show NEDDylation can promote degradation. We also examine how many of the in vitro studies measuring non-canonical NEDDylation were conducted and compare those conditions to those which may occur in vivo, such as cancer progression. It is likely that the conditions used to study non-canonical NEDDylation are similar to some types of cancers, such as glioblastoma, colon and rectal cancers, and lung adenocarcinomas. Although the full outcomes of non-canonical NEDDylation remain unknown, our review of the literature suggests that researchers keep an open mind to the situations where this modification occurs and determine the functional impacts of NEDD8-modification to the specific substrates which they study.
Keywords: NEDD8; post-translational modifications; ubiquitin-like proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function.Elife. 2017 May 5;6:e24325. doi: 10.7554/eLife.24325. Elife. 2017. PMID: 28475037 Free PMC article.
-
Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes.Biochem J. 2012 Feb 1;441(3):927-36. doi: 10.1042/BJ20111671. Biochem J. 2012. PMID: 22004789 Free PMC article.
-
NEDD8-its role in the regulation of Cullin-RING ligases.Curr Opin Plant Biol. 2018 Oct;45(Pt A):112-119. doi: 10.1016/j.pbi.2018.05.017. Epub 2018 Jun 15. Curr Opin Plant Biol. 2018. PMID: 29909289 Review.
-
Proteome-wide identification of NEDD8 modification sites reveals distinct proteomes for canonical and atypical NEDDylation.Cell Rep. 2021 Jan 19;34(3):108635. doi: 10.1016/j.celrep.2020.108635. Cell Rep. 2021. PMID: 33472076
-
Protein neddylation and its role in health and diseases.Signal Transduct Target Ther. 2024 Apr 5;9(1):85. doi: 10.1038/s41392-024-01800-9. Signal Transduct Target Ther. 2024. PMID: 38575611 Free PMC article. Review.
Cited by
-
Neddylation of Enterovirus 71 VP2 Protein Reduces Its Stability and Restricts Viral Replication.J Virol. 2022 May 25;96(10):e0059822. doi: 10.1128/jvi.00598-22. Epub 2022 May 5. J Virol. 2022. PMID: 35510863 Free PMC article.
-
Targeting the NEDP1 enzyme to ameliorate ALS phenotypes through stress granule disassembly.Sci Adv. 2023 Mar 31;9(13):eabq7585. doi: 10.1126/sciadv.abq7585. Epub 2023 Mar 31. Sci Adv. 2023. PMID: 37000881 Free PMC article.
-
Targeting NEDD8 suppresses surgical stress-facilitated metastasis of colon cancer via restraining regulatory T cells.Cell Death Dis. 2024 Jan 5;15(1):8. doi: 10.1038/s41419-023-06396-6. Cell Death Dis. 2024. PMID: 38177106 Free PMC article.
-
SUMOylation and NEDDylation in Primary and Metastatic Cancers to Bone.Front Cell Dev Biol. 2022 Apr 6;10:889002. doi: 10.3389/fcell.2022.889002. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35465332 Free PMC article. Review.
-
Neddylation signaling inactivation by tetracaine hydrochloride suppresses cell proliferation and alleviates vemurafenib-resistance of melanoma.Cell Biol Toxicol. 2024 Sep 19;40(1):81. doi: 10.1007/s10565-024-09916-y. Cell Biol Toxicol. 2024. PMID: 39297891 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

