CDK1-Mediated Phosphorylation of BAG3 Promotes Mitotic Cell Shape Remodeling and the Molecular Assembly of Mitotic p62 Bodies
- PMID: 34685619
- PMCID: PMC8534064
- DOI: 10.3390/cells10102638
CDK1-Mediated Phosphorylation of BAG3 Promotes Mitotic Cell Shape Remodeling and the Molecular Assembly of Mitotic p62 Bodies
Abstract
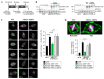
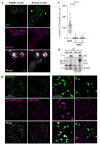
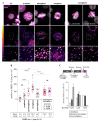
The cochaperone BCL2-associated athanogene 3 (BAG3), in complex with the heat shock protein HSPB8, facilitates mitotic rounding, spindle orientation, and proper abscission of daughter cells. BAG3 and HSPB8 mitotic functions implicate the sequestosome p62/SQSTM1, suggesting a role for protein quality control. However, the interplay between this chaperone-assisted pathway and the mitotic machinery is not known. Here, we show that BAG3 phosphorylation at the conserved T285 is regulated by CDK1 and activates its function in mitotic cell shape remodeling. BAG3 phosphorylation exhibited a high dynamic at mitotic entry and both a non-phosphorylatable BAG3T285A and a phosphomimetic BAG3T285D protein were unable to correct the mitotic defects in BAG3-depleted HeLa cells. We also demonstrate that BAG3 phosphorylation, HSPB8, and CDK1 activity modulate the molecular assembly of p62/SQSTM1 into mitotic bodies containing K63 polyubiquitinated chains. These findings suggest the existence of a mitotically regulated spatial quality control mechanism for the fidelity of cell shape remodeling in highly dividing cells.
Keywords: BAG3 1; CDK1 4; HSPB8 2; K63 polyubiquitin chain 8; actin 7; mitotic cell rounding 6; p62/SQSTM1 bodies 3; protein quality control 9; spindle positioning 5.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


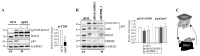
Similar articles
-
Chaperone-Assisted Mitotic Actin Remodeling by BAG3 and HSPB8 Involves the Deacetylase HDAC6 and Its Substrate Cortactin.Int J Mol Sci. 2020 Dec 25;22(1):142. doi: 10.3390/ijms22010142. Int J Mol Sci. 2020. PMID: 33375626 Free PMC article.
-
A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis.PLoS Genet. 2015 Oct 23;11(10):e1005582. doi: 10.1371/journal.pgen.1005582. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26496431 Free PMC article.
-
HSPB8 and BAG3 cooperate to promote spatial sequestration of ubiquitinated proteins and coordinate the cellular adaptive response to proteasome insufficiency.FASEB J. 2018 Jul;32(7):3518-3535. doi: 10.1096/fj.201700558RR. Epub 2018 Feb 5. FASEB J. 2018. PMID: 29405094
-
HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy.Autophagy. 2008 Feb;4(2):237-9. doi: 10.4161/auto.5407. Epub 2007 Dec 11. Autophagy. 2008. PMID: 18094623 Review.
-
The role of BAG3 in dilated cardiomyopathy and its association with Charcot-Marie-Tooth disease type 2.Acta Myol. 2022 Jun 30;41(2):59-75. doi: 10.36185/2532-1900-071. eCollection 2022 Jun. Acta Myol. 2022. PMID: 35832504 Free PMC article. Review.
Cited by
-
Identification of phosphatases that dephosphorylate the co-chaperone BAG3.Life Sci Alliance. 2024 Nov 19;8(2):e202402734. doi: 10.26508/lsa.202402734. Print 2025 Feb. Life Sci Alliance. 2024. PMID: 39562141 Free PMC article.
-
Genetics of BAG3: A Paradigm for Developing Precision Therapies for Dilated Cardiomyopathies.J Am Heart Assoc. 2022 Dec 6;11(23):e027373. doi: 10.1161/JAHA.122.027373. Epub 2022 Nov 16. J Am Heart Assoc. 2022. PMID: 36382946 Free PMC article. Review.
-
The Role of Small Heat Shock Proteins in Protein Misfolding Associated Motoneuron Diseases.Int J Mol Sci. 2022 Oct 4;23(19):11759. doi: 10.3390/ijms231911759. Int J Mol Sci. 2022. PMID: 36233058 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

