Long non-coding RNA SNHG17 promotes lung adenocarcinoma progression by targeting the microRNA-193a-5p/NETO2 axis
- PMID: 34671432
- PMCID: PMC8503812
- DOI: 10.3892/ol.2021.13079
Long non-coding RNA SNHG17 promotes lung adenocarcinoma progression by targeting the microRNA-193a-5p/NETO2 axis
Abstract
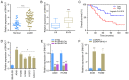
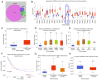
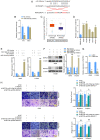
Long non-coding RNAs (lncRNAs) play vital roles in human cancers. It has been reported that lncRNA SNHG17 expression is dysregulated in different types of cancer and involved in cancer progression. However, the role of SNHG17 in lung adenocarcinoma (LUAD) remains unclear. The present study aimed to investigate the role of SNHG17 in LUAD. Reverse transcription-quantitative (RT-q) PCR analysis was performed to detect SNHG17 expression in LUAD tissues and cells. The effects of SNHG17 on cancer cell migration, invasion, proliferation and epithelial-to-mesenchymal transition (EMT) were assessed via Transwell, MTT and western blot assays, respectively. The interactions between SNHG17 and microRNA (miRNA/miR)-193a-5p, miR-193a-5p and neuropilin and tolloid-like 2 (NETO2) were assessed via the dual-luciferase reporter assay. NETO2 expression and its potential role in LUAD were analyzed via RT-qPCR analysis and the UALCAN database. The results demonstrated that SNHG17 expression was significantly upregulated in LUAD tissues and cells, and high SNHG17 expression was associated with tumor-node-metastasis stage and poor prognosis of patients with LUAD. SNHG17 knockdown inhibited cell migration, invasion, proliferation and the EMT process. In addition, the results revealed that SNHG17 functions as a competing endogenous RNA of miR-193a-5p. The results of the dual-luciferase reporter assay confirmed that miR-193a-5p can directly target SNHG17. NETO2 was also predicted as a target protein of miR-193a-5p, which was confirmed via the dual-luciferase reporter assay. The roles of NETO2 knockdown in cancer cells were rescued following transfection with miR-193a-5p inhibitor or overexpression of SNHG17. Notably, high NETO2 expression was associated with poor prognosis of patients with LUAD. Bioinformatics analysis demonstrated that the promoter methylation level of NETO2 decreased in LUAD. Taken together, the results of the present study suggest that SNHG17 expression is upregulated in LUAD tissues and cells, and SNHG17 exerts tumor promoting effect by targeting the miR-193a-5p/NETO2 axis.
Keywords: SNHG17; lung adenocarcinoma; microRNA-193a-5p; neuropilin and tolloid-like 2.
Copyright: © Zhang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
SNHG17 upregulates WLS expression to accelerate lung adenocarcinoma progression by sponging miR-485-5p.Biochem Biophys Res Commun. 2020 Dec 17;533(4):1435-1441. doi: 10.1016/j.bbrc.2020.09.130. Epub 2020 Oct 24. Biochem Biophys Res Commun. 2020. PMID: 33109341
-
The LncRNA NEAT1 Accelerates Lung Adenocarcinoma Deterioration and Binds to Mir-193a-3p as a Competitive Endogenous RNA.Cell Physiol Biochem. 2018;48(3):905-918. doi: 10.1159/000491958. Epub 2018 Jul 23. Cell Physiol Biochem. 2018. PMID: 30036873
-
LncRNA LINC00511 promotes COL1A1-mediated proliferation and metastasis by sponging miR-126-5p/miR-218-5p in lung adenocarcinoma.BMC Pulm Med. 2022 Jul 16;22(1):272. doi: 10.1186/s12890-022-02070-3. BMC Pulm Med. 2022. PMID: 35842617 Free PMC article.
-
LncRNA-AC009948.5 promotes invasion and metastasis of lung adenocarcinoma by binding to miR-186-5p.Front Oncol. 2022 Aug 19;12:949951. doi: 10.3389/fonc.2022.949951. eCollection 2022. Front Oncol. 2022. PMID: 36059662 Free PMC article.
-
LncRNA SGMS1-AS1 regulates lung adenocarcinoma cell proliferation, migration, invasion, and EMT progression via miR-106a-5p/MYLI9 axis.Thorac Cancer. 2021 Jul;12(14):2104-2112. doi: 10.1111/1759-7714.14043. Epub 2021 Jun 1. Thorac Cancer. 2021. PMID: 34061466 Free PMC article.
Cited by
-
The crucial roles of long noncoding RNA SNHGs in lung cancer.Clin Transl Oncol. 2022 Dec;24(12):2272-2284. doi: 10.1007/s12094-022-02909-5. Epub 2022 Aug 25. Clin Transl Oncol. 2022. PMID: 36008615 Review.
-
Integrative Analysis and Experimental Validation Indicated That SNHG17 Is a Prognostic Marker in Prostate Cancer and a Modulator of the Tumor Microenvironment via a Competitive Endogenous RNA Regulatory Network.Oxid Med Cell Longev. 2022 Jul 12;2022:1747604. doi: 10.1155/2022/1747604. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35864871 Free PMC article.
-
Long noncoding RNA SNHG17: a novel molecule in human cancers.Cancer Cell Int. 2022 Mar 5;22(1):104. doi: 10.1186/s12935-022-02529-7. Cancer Cell Int. 2022. PMID: 35248073 Free PMC article. Review.
-
[Research Progress of LncRNA SNHGs in Regulating the Biological Behavior of Non-small Cell Lung Cancer].Zhongguo Fei Ai Za Zhi. 2023 Nov 20;26(11):851-862. doi: 10.3779/j.issn.1009-3419.2023.102.41. Zhongguo Fei Ai Za Zhi. 2023. PMID: 38061887 Free PMC article. Review. Chinese.
-
Enabling factor for cancer hallmark acquisition: Small nucleolar RNA host gene 17.Front Oncol. 2022 Sep 14;12:974939. doi: 10.3389/fonc.2022.974939. eCollection 2022. Front Oncol. 2022. PMID: 36185210 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
