Proteomics and its applications in breast cancer
- PMID: 34659875
- PMCID: PMC8493401
Proteomics and its applications in breast cancer
Abstract
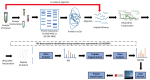
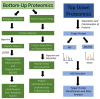
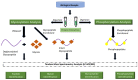
Breast cancer is an individually unique, multi-faceted and chameleonic disease, an eternal challenge for the new era of high-integrated precision diagnostic and personalized oncomedicine. Besides traditional single-omics fields (such as genomics, epigenomics, transcriptomics and metabolomics) and multi-omics contributions (proteogenomics, proteotranscriptomics or reproductomics), several new "-omics" approaches and exciting proteomics subfields are contributing to basic and advanced understanding of these "multiple diseases termed breast cancer": phenomics/cellomics, connectomics and interactomics, secretomics, matrisomics, exosomics, angiomics, chaperomics and epichaperomics, phosphoproteomics, ubiquitinomics, metalloproteomics, terminomics, degradomics and metadegradomics, adhesomics, stressomics, microbiomics, immunomics, salivaomics, materiomics and other biomics. Throughout the extremely complex neoplastic process, a Breast Cancer Cell Continuum Concept (BCCCC) has been modeled in this review as a spatio-temporal and holistic approach, as long as the breast cancer represents a complex cascade comprising successively integrated populations of heterogeneous tumor and cancer-associated cells, that reflect the carcinoma's progression from a "driving mutation" and formation of the breast primary tumor, toward the distant secondary tumors in different tissues and organs, via circulating tumor cell populations. This BCCCC is widely sustained by a Breast Cancer Proteomic Continuum Concept (BCPCC), where each phenotype of neoplastic and tumor-associated cells is characterized by a changing and adaptive proteomic profile detected in solid and liquid minimal invasive biopsies by complex proteomics approaches. Such a profile is created, beginning with the proteomic landscape of different neoplastic cell populations and cancer-associated cells, followed by subsequent analysis of protein biomarkers involved in epithelial-mesenchymal transition and intravasation, circulating tumor cell proteomics, and, finally, by protein biomarkers that highlight the extravasation and distant metastatic invasion. Proteomics technologies are producing important data in breast cancer diagnostic, prognostic, and predictive biomarkers discovery and validation, are detecting genetic aberrations at the proteome level, describing functional and regulatory pathways and emphasizing specific protein and peptide profiles in human tissues, biological fluids, cell lines and animal models. Also, proteomics can identify different breast cancer subtypes and specific protein and proteoform expression, can assess the efficacy of cancer therapies at cellular and tissular level and can even identify new therapeutic target proteins in clinical studies.
Keywords: Proteomics; biomarkers; breast cancer cell continuum concept; breast cancer proteomic continuum concept.
AJCR Copyright © 2021.
Conflict of interest statement
None.
Figures

Similar articles
-
Omics-Based Investigations of Breast Cancer.Molecules. 2023 Jun 14;28(12):4768. doi: 10.3390/molecules28124768. Molecules. 2023. PMID: 37375323 Free PMC article. Review.
-
"Omics" in pharmaceutical research: overview, applications, challenges, and future perspectives.Chin J Nat Med. 2015 Jan;13(1):3-21. doi: 10.1016/S1875-5364(15)60002-4. Chin J Nat Med. 2015. PMID: 25660284 Review.
-
Salivary Biomarkers in Breast Cancer: From Salivaomics to Salivaoncoomics.Front Biosci (Landmark Ed). 2024 Jul 17;29(7):253. doi: 10.31083/j.fbl2907253. Front Biosci (Landmark Ed). 2024. PMID: 39082349 Review.
-
[OMICS and biomarkers of glial tumors].Rev Neurol (Paris). 2011 Oct;167(10):691-8. doi: 10.1016/j.neurol.2011.07.007. Epub 2011 Sep 1. Rev Neurol (Paris). 2011. PMID: 21889780 Review. French.
-
High-throughput «Omics» technologies: New tools for the study of triple-negative breast cancer.Cancer Lett. 2016 Nov 1;382(1):77-85. doi: 10.1016/j.canlet.2016.03.001. Epub 2016 Mar 7. Cancer Lett. 2016. PMID: 26965997 Review.
Cited by
-
Longitudinal Serum Protein Analysis of Women with a High Risk of Developing Breast Cancer Reveals Large Interpatient Versus Small Intrapatient Variations: First Results from the TESTBREAST Study.Int J Mol Sci. 2022 Oct 17;23(20):12399. doi: 10.3390/ijms232012399. Int J Mol Sci. 2022. PMID: 36293255 Free PMC article.
-
Proteomics Analysis of Lymphoblastoid Cell Lines from Patients with Amyotrophic Lateral Sclerosis.Molecules. 2023 Feb 21;28(5):2014. doi: 10.3390/molecules28052014. Molecules. 2023. PMID: 36903260 Free PMC article.
-
Breast Sarcomas-How Different Are They from Breast Carcinomas? Clinical, Pathological, Imaging and Treatment Insights.Diagnostics (Basel). 2023 Apr 7;13(8):1370. doi: 10.3390/diagnostics13081370. Diagnostics (Basel). 2023. PMID: 37189471 Free PMC article.
-
Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer.Proteomes. 2022 Oct 21;10(4):35. doi: 10.3390/proteomes10040035. Proteomes. 2022. PMID: 36278695 Free PMC article. Review.
-
Delineating intra-tumoral heterogeneity and tumor evolution in breast cancer using precision-based approaches.Front Genet. 2023 Aug 17;14:1087432. doi: 10.3389/fgene.2023.1087432. eCollection 2023. Front Genet. 2023. PMID: 37662839 Free PMC article. Review.
References
-
- Elzamly S, Badri N, Padilla O, Dwivedi AK, Alvarado LA, Hamilton M, Diab N, Rock C, Elfar A, Teleb M, Sanchez L, Nahleh Z. Epithelial-mesenchymal transition markers in breast cancer and pathological responseafter neoadjuvant chemotherapy. Breast Cancer (Auckl) 2018;12:1178223418788074. - PMC - PubMed
-
- Coleman WB. Next generation breast cancer omics. Am J Pathol. 2017;187:2130–2132. - PubMed
-
- Davatzikos C, Rathore S, Bakas S, Pati S, Bergman M, Kalarot R, Sridharan P, Gastounioti A, Jahani N, Cohen E, Akbari H, Tunc B, Doshi J, Parker D, Hsieh M, Sotiras A, Li H, Ou Y, Doot RK, Bilello M, Fan Y, Shinohara RT, Yushkevich P, Verma R, Kontos D. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J Med Imaging (Bellingham) 2018;5:011018. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
