A sticky situation: regulation and function of protein palmitoylation with a spotlight on the axon and axon initial segment
- PMID: 34659801
- PMCID: PMC8495546
- DOI: 10.1042/NS20210005
A sticky situation: regulation and function of protein palmitoylation with a spotlight on the axon and axon initial segment
Abstract
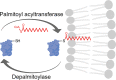
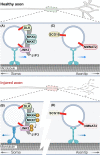
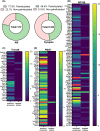
In neurons, the axon and axon initial segment (AIS) are critical structures for action potential initiation and propagation. Their formation and function rely on tight compartmentalisation, a process where specific proteins are trafficked to and retained at distinct subcellular locations. One mechanism which regulates protein trafficking and association with lipid membranes is the modification of protein cysteine residues with the 16-carbon palmitic acid, known as S-acylation or palmitoylation. Palmitoylation, akin to phosphorylation, is reversible, with palmitate cycling being mediated by substrate-specific enzymes. Palmitoylation is well-known to be highly prevalent among neuronal proteins and is well studied in the context of the synapse. Comparatively, how palmitoylation regulates trafficking and clustering of axonal and AIS proteins remains less understood. This review provides an overview of the current understanding of the biochemical regulation of palmitoylation, its involvement in various neurological diseases, and the most up-to-date perspective on axonal palmitoylation. Through a palmitoylation analysis of the AIS proteome, we also report that an overwhelming proportion of AIS proteins are likely palmitoylated. Overall, our review and analysis confirm a central role for palmitoylation in the formation and function of the axon and AIS and provide a resource for further exploration of palmitoylation-dependent protein targeting to and function at the AIS.
Keywords: S-acylation; acyl protein thioesterase; axon; axon initial segment; palmitoyl acyltransferase; palmitoylation.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
Lost in traffic: consequences of altered palmitoylation in neurodegeneration.Front Physiol. 2023 May 30;14:1166125. doi: 10.3389/fphys.2023.1166125. eCollection 2023. Front Physiol. 2023. PMID: 37324388 Free PMC article. Review.
-
Detection of protein palmitoylation in cultured hippocampal neurons by immunoprecipitation and acyl-biotin exchange (ABE).J Vis Exp. 2013 Feb 18;(72):50031. doi: 10.3791/50031. J Vis Exp. 2013. PMID: 23438969 Free PMC article.
-
Protein Palmitoylation by DHHC Protein Family.In: Kittler JT, Moss SJ, editors. The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology. Boca Raton (FL): CRC Press/Taylor & Francis; 2006. Chapter 5. In: Kittler JT, Moss SJ, editors. The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology. Boca Raton (FL): CRC Press/Taylor & Francis; 2006. Chapter 5. PMID: 21204476 Free Books & Documents. Review.
-
Emerging roles for protein S-palmitoylation in Toxoplasma biology.Int J Parasitol. 2014 Feb;44(2):121-31. doi: 10.1016/j.ijpara.2013.09.004. Epub 2013 Nov 1. Int J Parasitol. 2014. PMID: 24184909 Review.
-
Targeting of Specialized Metabolites Biosynthetic Enzymes to Membranes and Vesicles by Posttranslational Palmitoylation: A Mechanism of Non-Conventional Traffic and Secretion of Fungal Metabolites.Int J Mol Sci. 2024 Jan 19;25(2):1224. doi: 10.3390/ijms25021224. Int J Mol Sci. 2024. PMID: 38279221 Free PMC article. Review.
Cited by
-
Full-length huntingtin is palmitoylated at multiple sites and post-translationally myristoylated following caspase-cleavage.Front Physiol. 2023 Jan 13;14:1086112. doi: 10.3389/fphys.2023.1086112. eCollection 2023. Front Physiol. 2023. PMID: 36711022 Free PMC article.
-
Mechanisms and functions of protein S-acylation.Nat Rev Mol Cell Biol. 2024 Jun;25(6):488-509. doi: 10.1038/s41580-024-00700-8. Epub 2024 Feb 14. Nat Rev Mol Cell Biol. 2024. PMID: 38355760 Review.
-
Lost in traffic: consequences of altered palmitoylation in neurodegeneration.Front Physiol. 2023 May 30;14:1166125. doi: 10.3389/fphys.2023.1166125. eCollection 2023. Front Physiol. 2023. PMID: 37324388 Free PMC article. Review.
-
Altered Protein Palmitoylation as Disease Mechanism in Neurodegenerative Disorders.J Neurosci. 2024 Oct 2;44(40):e1225242024. doi: 10.1523/JNEUROSCI.1225-24.2024. J Neurosci. 2024. PMID: 39358031 Review.
References
-
- Fukata Y. and Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175, https://www.nature.com/articles/nrn2788 10.1038/nrn2788 - DOI - PubMed
-
- Sutton L.M., Sanders S.S., Butland S.L., Singaraja R.R., Franciosi S., Southwell A.L.et al. . (2013) Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease. Hum. Mol. Genet. 22, 452–465, http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id... 10.1093/hmg/dds441 - DOI - PubMed
-
- Singaraja R.R., Huang K., Sanders S.S., Milnerwood A.J., Hines R., Lerch J.P.et al. . (2011) Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum. Mol. Genet. 20, 3899–3909, http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id... 10.1093/hmg/ddr308 - DOI - PMC - PubMed
-
- Raymond F.L., Tarpey P.S., Edkins S., Tofts C., O’Meara S., Teague J.et al. . (2007) Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid Habitus. Am. J. Hum. Genet. 80, 982–987, http://linkinghub.elsevier.com/retrieve/pii/S0002929707609549 10.1086/513609 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

