Transcriptomic Signatures Associated With Regional Cortical Thickness Changes in Parkinson's Disease
- PMID: 34658772
- PMCID: PMC8519261
- DOI: 10.3389/fnins.2021.733501
Transcriptomic Signatures Associated With Regional Cortical Thickness Changes in Parkinson's Disease
Abstract
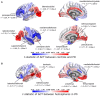
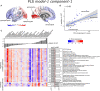
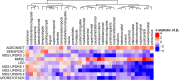
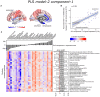
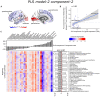
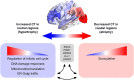
Cortical atrophy is a common manifestation in Parkinson's disease (PD), particularly in advanced stages of the disease. To elucidate the molecular underpinnings of cortical thickness changes in PD, we performed an integrated analysis of brain-wide healthy transcriptomic data from the Allen Human Brain Atlas and patterns of cortical thickness based on T1-weighted anatomical MRI data of 149 PD patients and 369 controls. For this purpose, we used partial least squares regression to identify gene expression patterns correlated with cortical thickness changes. In addition, we identified gene expression patterns underlying the relationship between cortical thickness and clinical domains of PD. Our results show that genes whose expression in the healthy brain is associated with cortical thickness changes in PD are enriched in biological pathways related to sumoylation, regulation of mitotic cell cycle, mitochondrial translation, DNA damage responses, and ER-Golgi traffic. The associated pathways were highly related to each other and all belong to cellular maintenance mechanisms. The expression of genes within most pathways was negatively correlated with cortical thickness changes, showing higher expression in regions associated with decreased cortical thickness (atrophy). On the other hand, sumoylation pathways were positively correlated with cortical thickness changes, showing higher expression in regions with increased cortical thickness (hypertrophy). Our findings suggest that alterations in the balanced interplay of these mechanisms play a role in changes of cortical thickness in PD and possibly influence motor and cognitive functions.
Keywords: cortical thickness; gene expression analysis; imaging-genetics; neurodegenerative diseases; neuroimaging data.
Copyright © 2021 Keo, Dzyubachyk, van der Grond, van Hilten, Reinders and Mahfouz.
Conflict of interest statement
The authors declare that this study received funding from AbbVie, Hoffmann-La-Roche, and Lundbeck. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Figures
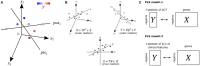

Similar articles
-
Morphometric similarity differences in drug-naive Parkinson's disease correlate with transcriptomic signatures.CNS Neurosci Ther. 2024 Mar;30(3):e14680. doi: 10.1111/cns.14680. CNS Neurosci Ther. 2024. PMID: 38529533 Free PMC article.
-
Motor symptoms in Parkinson's disease are related to the interplay between cortical curvature and thickness.Neuroimage Clin. 2023;37:103300. doi: 10.1016/j.nicl.2022.103300. Epub 2022 Dec 21. Neuroimage Clin. 2023. PMID: 36580712 Free PMC article.
-
Cortical atrophy is associated with cognitive impairment in Parkinson's disease: a combined analysis of cortical thickness and functional connectivity.Brain Imaging Behav. 2022 Dec;16(6):2586-2600. doi: 10.1007/s11682-022-00714-w. Epub 2022 Aug 31. Brain Imaging Behav. 2022. PMID: 36044168
-
Longitudinal brain atrophy distribution in advanced Parkinson's disease: What makes the difference in "cognitive status" converters?Hum Brain Mapp. 2020 Apr 15;41(6):1416-1434. doi: 10.1002/hbm.24884. Epub 2019 Dec 2. Hum Brain Mapp. 2020. PMID: 31789477 Free PMC article.
-
Microstructural but not macrostructural cortical degeneration occurs in Parkinson's disease with mild cognitive impairment.NPJ Parkinsons Dis. 2022 Nov 9;8(1):151. doi: 10.1038/s41531-022-00416-6. NPJ Parkinsons Dis. 2022. PMID: 36351910 Free PMC article. Review.
Cited by
-
Alzheimer's Disease-Related Genes Identified by Linking Spatial Patterns of Pathology and Gene Expression.Front Neurosci. 2022 Jun 14;16:908650. doi: 10.3389/fnins.2022.908650. eCollection 2022. Front Neurosci. 2022. PMID: 35774552 Free PMC article.
References
LinkOut - more resources
Full Text Sources

