Ssl2/TFIIH function in transcription start site scanning by RNA polymerase II in Saccharomyces cerevisiae
- PMID: 34652274
- PMCID: PMC8589449
- DOI: 10.7554/eLife.71013
Ssl2/TFIIH function in transcription start site scanning by RNA polymerase II in Saccharomyces cerevisiae
Abstract
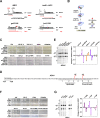
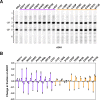
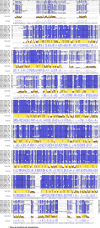
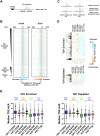
In Saccharomyces cerevisiae, RNA polymerase II (Pol II) selects transcription start sites (TSSs) by a unidirectional scanning process. During scanning, a preinitiation complex (PIC) assembled at an upstream core promoter initiates at select positions within a window ~40-120 bp downstream. Several lines of evidence indicate that Ssl2, the yeast homolog of XPB and an essential and conserved subunit of the general transcription factor (GTF) TFIIH, drives scanning through its DNA-dependent ATPase activity, therefore potentially controlling both scanning rate and scanning extent (processivity). To address questions of how Ssl2 functions in promoter scanning and interacts with other initiation activities, we leveraged distinct initiation-sensitive reporters to identify novel ssl2 alleles. These ssl2 alleles, many of which alter residues conserved from yeast to human, confer either upstream or downstream TSS shifts at the model promoter ADH1 and genome-wide. Specifically, tested ssl2 alleles alter TSS selection by increasing or narrowing the distribution of TSSs used at individual promoters. Genetic interactions of ssl2 alleles with other initiation factors are consistent with ssl2 allele classes functioning through increasing or decreasing scanning processivity but not necessarily scanning rate. These alleles underpin a residue interaction network that likely modulates Ssl2 activity and TFIIH function in promoter scanning. We propose that the outcome of promoter scanning is determined by two functional networks, the first being Pol II activity and factors that modulate it to determine initiation efficiency within a scanning window, and the second being Ssl2/TFIIH and factors that modulate scanning processivity to determine the width of the scanning widow.
Keywords: RNA polymerase II; S. cerevisiae; general transcription factors; genetics; genomics; transcription initiation; transcription start site; yeast.
Plain language summary
In eukaryotic organisms such as yeast, the process of converting genes into proteins begins with the transcription of DNA sequences into mRNA molecules. An enzyme called RNA Polymerase II (Pol II) is responsible for creating new strands of mRNA, but a variety of other so called transcription factors is also needed to kickstart the transcription process. These transcription factors are delivered to genes, where they attach to specific sequences, or promoters, which sit at the beginning of each gene. Once these transcription factors are in place, the double stranded DNA is unzipped to provide access to the DNA that will serve as the template for transcription. In budding yeast, Pol II and another specific transcription factor, known as TFIIH, work together to scan these promoter sequences to find the appropriate start sites of mRNA synthesis. However, several aspects of this process, such as how TFIIH works in promoter scanning, how far its scanning functions can extend, and how its activity is controlled, are currently poorly understood. Zhao et al. have investigated these questions in budding yeast. Using a range of genetic and genomic techniques, Zhao et al. found that certain sections of TFIIH were involved in choosing specific transcription start sites of mRNA synthesis during promoter scanning. These sections were identical in different eukaryotic organisms from yeast to humans, suggesting that these regions may be important for tuning or controlling the activity of TFIIH. Moreover, in yeast, the activity of TFIIH determines how far the scanning unit was able to move along the promoter DNA. Finally, Zhao et al. found that the initiation by promoter scanning was regulated by two distinct networks. The first network controlled how well mRNA synthesis could be initiated at individual transcription start sites; and the second network – driven by TFIIH – controlled which promoter sequences could be scanned to initiate transcription. This research provides an in-depth look into the early steps of the process of converting DNA into mRNA. The biological machinery used to initiate and control this action is highly conserved between yeast and humans, suggesting that the mechanisms for controlling the activity of these factors could be similar, even if their initiation processes may differ.
© 2021, Zhao et al.
Conflict of interest statement
TZ, IV, WL, SB, BN, CK No competing interests declared, BP BFP has a financial interest in Peconic, LLC, which utilizes the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research.
Figures












Similar articles
-
The Role of XPB/Ssl2 dsDNA Translocase Processivity in Transcription Start-site Scanning.J Mol Biol. 2021 Jul 9;433(14):166813. doi: 10.1016/j.jmb.2021.166813. Epub 2021 Jan 13. J Mol Biol. 2021. PMID: 33453189 Free PMC article.
-
Systematic mutagenesis of TFIIH subunit p52/Tfb2 identifies residues required for XPB/Ssl2 subunit function and genetic interactions with TFB6.J Biol Chem. 2022 Oct;298(10):102433. doi: 10.1016/j.jbc.2022.102433. Epub 2022 Aug 28. J Biol Chem. 2022. PMID: 36041630 Free PMC article.
-
Mechanism of start site selection by RNA polymerase II: interplay between TFIIB and Ssl2/XPB helicase subunit of TFIIH.J Biol Chem. 2012 Jan 2;287(1):557-567. doi: 10.1074/jbc.M111.281576. Epub 2011 Nov 11. J Biol Chem. 2012. PMID: 22081613 Free PMC article.
-
Single-molecule approach for studying RNAP II transcription initiation using magnetic tweezers.Methods. 2019 Apr 15;159-160:35-44. doi: 10.1016/j.ymeth.2019.03.010. Epub 2019 Mar 18. Methods. 2019. PMID: 30898685 Free PMC article. Review.
-
Interplay between the transcription preinitiation complex and the +1 nucleosome.Trends Biochem Sci. 2024 Feb;49(2):145-155. doi: 10.1016/j.tibs.2023.12.001. Epub 2024 Jan 13. Trends Biochem Sci. 2024. PMID: 38218671 Review.
Cited by
-
Disordered C-terminal domain drives spatiotemporal confinement of RNAPII to enhance search for chromatin targets.Nat Cell Biol. 2024 Apr;26(4):581-592. doi: 10.1038/s41556-024-01382-2. Epub 2024 Mar 28. Nat Cell Biol. 2024. PMID: 38548891 Free PMC article.
-
Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.Mol Cell. 2022 Feb 3;82(3):660-676.e9. doi: 10.1016/j.molcel.2021.12.020. Epub 2022 Jan 19. Mol Cell. 2022. PMID: 35051353 Free PMC article.
-
Quantitative analysis of transcription start site selection reveals control by DNA sequence, RNA polymerase II activity and NTP levels.Nat Struct Mol Biol. 2024 Jan;31(1):190-202. doi: 10.1038/s41594-023-01171-9. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177677 Free PMC article.
-
Live-cell analysis of IMPDH protein levels during yeast colony growth provides insights into the regulation of GTP synthesis.mBio. 2024 Aug 14;15(8):e0102124. doi: 10.1128/mbio.01021-24. Epub 2024 Jun 28. mBio. 2024. PMID: 38940616 Free PMC article.
-
Disordered C-terminal domain drives spatiotemporal confinement of RNAPII to enhance search for chromatin targets.bioRxiv [Preprint]. 2023 Nov 30:2023.07.31.551302. doi: 10.1101/2023.07.31.551302. bioRxiv. 2023. Update in: Nat Cell Biol. 2024 Apr;26(4):581-592. doi: 10.1038/s41556-024-01382-2 PMID: 37577667 Free PMC article. Updated. Preprint.
References
-
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2005.
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRP295731
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

