Impact of Helicobacter pylori eradication on the gastric microbiome
- PMID: 34645495
- PMCID: PMC8513236
- DOI: 10.1186/s13099-021-00460-2
Impact of Helicobacter pylori eradication on the gastric microbiome
Abstract
Background: Helicobacter pylori (Hp) eradication has been used for many years. Yet, the impact of this eradication on the normal gastric microflora is not well understood. In this study, we explored the effect of eradication on the stomach microbial community and its recovery after successful Hp eradication.
Methods: Among the 89 included patients, 23, 17, 40, and 9 were included in the Hp-negative, Hp-positive, successful eradication, and failed eradication groups, respectively. Four subgroups were further determined according to disease status (Hp-negative chronic gastritis [N-CG], Hp-negative atrophic gastritis [N-AG], successful-eradication chronic gastritis [SE-CG], and atrophic gastritis with successful eradication [SE-AG]). During the endoscopic examination, one piece of gastric mucosa tissue was obtained from the lesser curvature side of the gastric antrum and gastric corpus, respectively. In addition, 16S rDNA gene sequencing was used to analyze the gastric mucosal microbiome.
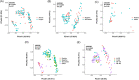
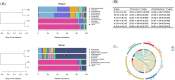
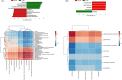
Results: In the Hp-negative group, the gastric microbiota was dominated by five phyla: Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, and Fusobacteria. After successfully eradicating Hp, the bacterial flora in the stomach recovered to a considerable extent. In the failed eradication group, the flora was similar to the flora in Hp-positive subjects based on the alpha and beta diversities. Among the groups, Curvibacter and Acinetobacter were enriched in the presence of Hp (i.e., failed eradication and Hp-positive groups), suggesting that these two genera could be used as biomarkers in the symbiotic flora in the presence of Hp. SE-CG was characterized by an increase in Firmicutes taxa and a decrease in Proteobacteria taxa compared with N-CG. SE-AG was characterized by a decrease in Firmicutes relative to N-AG. Finally, no differences were found in the pairwise comparisons of nitrate and nitrite reductase functions of the microflora among the four subgroups.
Conclusions: After Hp infection, the diversity and relative abundance of gastric microflora were significantly decreased. Yet, gastric microbiota could be partially restored to the Hp-negative status after eradication. Still, this effect was incomplete and might contribute to the long-term risks.
Keywords: 16S rDNA gene sequencing; Atrophic gastritis; Eradication therapy; Gastric microflora; Helicobacter pylori.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Gene expression of ornithine decarboxylase, cyclooxygenase-2, and gastrin in atrophic gastric mucosa infected with Helicobacter pylori before and after eradication therapy.Dig Dis Sci. 2003 Jan;48(1):36-46. doi: 10.1023/a:1021774029089. Dig Dis Sci. 2003. PMID: 12645788
-
Changes in Gastric Corpus Microbiota With Age and After Helicobacter pylori Eradication: A Long-Term Follow-Up Study.Front Microbiol. 2021 Feb 9;11:621879. doi: 10.3389/fmicb.2020.621879. eCollection 2020. Front Microbiol. 2021. PMID: 33633697 Free PMC article.
-
Oxidative damage of the gastric mucosa in Helicobacter pylori positive chronic atrophic and nonatrophic gastritis, before and after eradication.Helicobacter. 2003;8(5):503-12. doi: 10.1046/j.1523-5378.2003.00172.x. Helicobacter. 2003. PMID: 14535997
-
Treatment of Helicobacter pylori infection in atrophic gastritis.World J Gastroenterol. 2018 Jun 14;24(22):2373-2380. doi: 10.3748/wjg.v24.i22.2373. World J Gastroenterol. 2018. PMID: 29904244 Free PMC article. Review.
-
Helicobacter pylori, gastric microbiota and gastric cancer relationship: Unrolling the tangle.World J Gastrointest Oncol. 2022 May 15;14(5):959-972. doi: 10.4251/wjgo.v14.i5.959. World J Gastrointest Oncol. 2022. PMID: 35646287 Free PMC article. Review.
Cited by
-
Intestinal Macrophage Autophagy and its Pharmacological Application in Inflammatory Bowel Disease.Front Pharmacol. 2021 Nov 24;12:803686. doi: 10.3389/fphar.2021.803686. eCollection 2021. Front Pharmacol. 2021. PMID: 34899362 Free PMC article. Review.
-
Effect of Helicobacter Pylori Eradication on Human Gastric Microbiota: A Systematic Review and Meta-Analysis.Front Cell Infect Microbiol. 2022 May 4;12:899248. doi: 10.3389/fcimb.2022.899248. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35601105 Free PMC article.
-
Gastric microbiota in gastric cancer: Different roles of Helicobacter pylori and other microbes.Front Cell Infect Microbiol. 2023 Jan 10;12:1105811. doi: 10.3389/fcimb.2022.1105811. eCollection 2022. Front Cell Infect Microbiol. 2023. PMID: 36704105 Free PMC article. Review.
-
Bile reflux alters the profile of the gastric mucosa microbiota.Front Cell Infect Microbiol. 2022 Sep 9;12:940687. doi: 10.3389/fcimb.2022.940687. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36159635 Free PMC article.
-
Dual therapy for Helicobacter pylori infection.Chin Med J (Engl). 2023 Jan 5;136(1):13-23. doi: 10.1097/CM9.0000000000002565. Chin Med J (Engl). 2023. PMID: 36805362 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

