Endoplasmic Reticulum Chaperones in Viral Infection: Therapeutic Perspectives
- PMID: 34643441
- PMCID: PMC8515930
- DOI: 10.1128/MMBR.00035-21
Endoplasmic Reticulum Chaperones in Viral Infection: Therapeutic Perspectives
Abstract
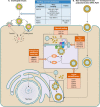
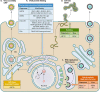
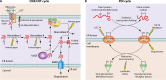
Viruses are intracellular parasites that subvert the functions of their host cells to accomplish their infection cycle. The endoplasmic reticulum (ER)-residing chaperone proteins are central for the achievement of different steps of the viral cycle, from entry and replication to assembly and exit. The most abundant ER chaperones are GRP78 (78-kDa glucose-regulated protein), GRP94 (94-kDa glucose-regulated protein), the carbohydrate or lectin-like chaperones calnexin (CNX) and calreticulin (CRT), the protein disulfide isomerases (PDIs), and the DNAJ chaperones. This review will focus on the pleiotropic roles of ER chaperones during viral infection. We will cover their essential role in the folding and quality control of viral proteins, notably viral glycoproteins which play a major role in host cell infection. We will also describe how viruses co-opt ER chaperones at various steps of their infectious cycle but also in order to evade immune responses and avoid apoptosis. Finally, we will discuss the different molecules targeting these chaperones and the perspectives in the development of broad-spectrum antiviral drugs.
Keywords: DNAJ; ER chaperone; GRP78; GRP94; calnexin; calreticulin; protein disulfide isomerase; viral infection.
Figures

Similar articles
-
Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins.J Virol. 1998 May;72(5):3851-8. doi: 10.1128/JVI.72.5.3851-3858.1998. J Virol. 1998. PMID: 9557669 Free PMC article.
-
Dimerization of ER-resident molecular chaperones mediated by ERp29.Biochem Biophys Res Commun. 2021 Jan 15;536:52-58. doi: 10.1016/j.bbrc.2020.12.023. Epub 2020 Dec 25. Biochem Biophys Res Commun. 2021. PMID: 33360823
-
Calnexin cycle - structural features of the ER chaperone system.FEBS J. 2020 Oct;287(20):4322-4340. doi: 10.1111/febs.15330. Epub 2020 Apr 27. FEBS J. 2020. PMID: 32285592 Free PMC article. Review.
-
Endoplasmic reticulum chaperones are involved in the morphogenesis of rotavirus infectious particles.J Virol. 2008 Jun;82(11):5368-80. doi: 10.1128/JVI.02751-07. Epub 2008 Apr 2. J Virol. 2008. PMID: 18385250 Free PMC article.
-
In vitro assays of the functions of calnexin and calreticulin, lectin chaperones of the endoplasmic reticulum.Methods Mol Biol. 2006;347:331-42. doi: 10.1385/1-59745-167-3:331. Methods Mol Biol. 2006. PMID: 17072021 Review.
Cited by
-
GRP78 Inhibitor YUM70 Suppresses SARS-CoV-2 Viral Entry, Spike Protein Production and Ameliorates Lung Damage.Viruses. 2023 May 6;15(5):1118. doi: 10.3390/v15051118. Viruses. 2023. PMID: 37243204 Free PMC article.
-
BiP/GRP78 is a pro-viral factor for diverse dsDNA viruses that promotes the survival and proliferation of cells upon KSHV infection.PLoS Pathog. 2024 Oct 29;20(10):e1012660. doi: 10.1371/journal.ppat.1012660. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39471213 Free PMC article.
-
Characterization and subcellular localization of Alongshan virus proteins.Front Microbiol. 2022 Sep 27;13:1000322. doi: 10.3389/fmicb.2022.1000322. eCollection 2022. Front Microbiol. 2022. PMID: 36238596 Free PMC article.
-
The stress-inducible ER chaperone GRP78/BiP is upregulated during SARS-CoV-2 infection and acts as a pro-viral protein.Nat Commun. 2022 Nov 14;13(1):6551. doi: 10.1038/s41467-022-34065-3. Nat Commun. 2022. PMID: 36376289 Free PMC article. No abstract available.
-
DNAJA4 Promotes the Replication of the Chinese Giant Salamander Iridovirus.Genes (Basel). 2022 Dec 24;14(1):58. doi: 10.3390/genes14010058. Genes (Basel). 2022. PMID: 36672799 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

