Thousands of Qatari genomes inform human migration history and improve imputation of Arab haplotypes
- PMID: 34642339
- PMCID: PMC8511259
- DOI: 10.1038/s41467-021-25287-y
Thousands of Qatari genomes inform human migration history and improve imputation of Arab haplotypes
Abstract
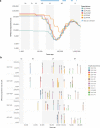
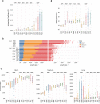
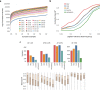
Arab populations are largely understudied, notably their genetic structure and history. Here we present an in-depth analysis of 6,218 whole genomes from Qatar, revealing extensive diversity as well as genetic ancestries representing the main founding Arab genealogical lineages of Qahtanite (Peninsular Arabs) and Adnanite (General Arabs and West Eurasian Arabs). We find that Peninsular Arabs are the closest relatives of ancient hunter-gatherers and Neolithic farmers from the Levant, and that founder Arab populations experienced multiple splitting events 12-20 kya, consistent with the aridification of Arabia and farming in the Levant, giving rise to settler and nomadic communities. In terms of recent genetic flow, we show that these ancestries contributed significantly to European, South Asian as well as South American populations, likely as a result of Islamic expansion over the past 1400 years. Notably, we characterize a large cohort of men with the ChrY J1a2b haplogroup (n = 1,491), identifying 29 unique sub-haplogroups. Finally, we leverage genotype novelty to build a reference panel of 12,432 haplotypes, demonstrating improved genotype imputation for both rare and common alleles in Arabs and the wider Middle East.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations.Genome Res. 2016 Feb;26(2):151-62. doi: 10.1101/gr.191478.115. Epub 2016 Jan 4. Genome Res. 2016. PMID: 26728717 Free PMC article.
-
Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago.BMC Evol Biol. 2018 Jun 19;18(1):98. doi: 10.1186/s12862-018-1211-4. BMC Evol Biol. 2018. PMID: 29921229 Free PMC article.
-
Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe.Proc Natl Acad Sci U S A. 2018 Jun 26;115(26):6774-6779. doi: 10.1073/pnas.1800851115. Epub 2018 Jun 12. Proc Natl Acad Sci U S A. 2018. PMID: 29895688 Free PMC article.
-
Population history of North Africa based on modern and ancient genomes.Hum Mol Genet. 2021 Apr 26;30(R1):R17-R23. doi: 10.1093/hmg/ddaa261. Hum Mol Genet. 2021. PMID: 33284971 Review.
-
Generalizability of GWA-Identified Genetic Risk Variants for Metabolic Traits to Populations from the Arabian Peninsula.Genes (Basel). 2021 Oct 18;12(10):1637. doi: 10.3390/genes12101637. Genes (Basel). 2021. PMID: 34681031 Free PMC article. Review.
Cited by
-
QPGx-CARES: Qatar pharmacogenetics clinical applications and research enhancement strategies.Clin Transl Sci. 2024 Jun;17(6):e13800. doi: 10.1111/cts.13800. Clin Transl Sci. 2024. PMID: 38818903 Free PMC article. Review.
-
Founder mutations and rare disease in the Arab world.Dis Model Mech. 2024 Jun 1;17(6):dmm050715. doi: 10.1242/dmm.050715. Epub 2024 Jun 26. Dis Model Mech. 2024. PMID: 38922202 Free PMC article. Review.
-
The impact of consanguinity on the design of iPSC banks.Stem Cell Reports. 2024 Aug 13;19(8):1043-1047. doi: 10.1016/j.stemcr.2024.06.004. Epub 2024 Jul 11. Stem Cell Reports. 2024. PMID: 38996491 Free PMC article.
-
Imputation Accuracy Across Global Human Populations.bioRxiv [Preprint]. 2023 Oct 26:2023.05.22.541241. doi: 10.1101/2023.05.22.541241. bioRxiv. 2023. Update in: Am J Hum Genet. 2024 May 2;111(5):979-989. doi: 10.1016/j.ajhg.2024.03.011. PMID: 37292811 Free PMC article. Updated. Preprint.
-
Poking COVID-19: Insights on Genomic Constraints among Immune-Related Genes between Qatari and Italian Populations.Genes (Basel). 2021 Nov 22;12(11):1842. doi: 10.3390/genes12111842. Genes (Basel). 2021. PMID: 34828448 Free PMC article.
References
-
- Al-Gazali L, Hamamy H, Al-Arrayad S. Genetic disorders in the Arab world. Br. Med. J. 2006;333:831–834. doi: 10.1136/bmj.38982.704931.AE. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

