Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity
- PMID: 34639110
- PMCID: PMC8509813
- DOI: 10.3390/ijms221910771
Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity
Abstract
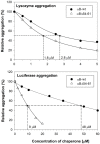
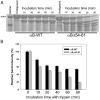
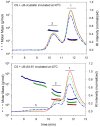
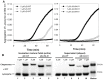
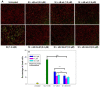
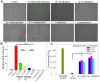
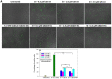
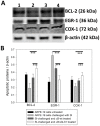
Previously, we showed that the removal of the 54-61 residues from αB-crystallin (αBΔ54-61) results in a fifty percent reduction in the oligomeric mass and a ten-fold increase in chaperone-like activity. In this study, we investigated the oligomeric organization changes in the deletion mutant contributing to the increased chaperone activity and evaluated the cytoprotection properties of the mutant protein using ARPE-19 cells. Trypsin digestion studies revealed that additional tryptic cleavage sites become susceptible in the deletion mutant than in the wild-type protein, suggesting a different subunit organization in the oligomer of the mutant protein. Static and dynamic light scattering analyses of chaperone-substrate complexes showed that the deletion mutant has more significant interaction with the substrates than wild-type protein, resulting in increased binding of the unfolding proteins. Cytotoxicity studies carried out with ARPE-19 cells showed an enhancement in anti-apoptotic activity in αBΔ54-61 as compared with the wild-type protein. The improved anti-apoptotic activity of the mutant is also supported by reduced caspase activation and normalization of the apoptotic cascade components level in cells treated with the deletion mutant. Our study suggests that altered oligomeric assembly with increased substrate affinity could be the basis for the enhanced chaperone function of the αBΔ54-61 protein.
Keywords: aggregation; apoptosis; cataract; chaperone; interactions; mutant; oligomerization; αB-crystallin.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Deletion of Specific Conserved Motifs from the N-Terminal Domain of αB-Crystallin Results in the Activation of Chaperone Functions.Int J Mol Sci. 2022 Jan 20;23(3):1099. doi: 10.3390/ijms23031099. Int J Mol Sci. 2022. PMID: 35163023 Free PMC article.
-
Deletion of (54)FLRAPSWF(61) residues decreases the oligomeric size and enhances the chaperone function of alphaB-crystallin.Biochemistry. 2009 Jun 16;48(23):5066-73. doi: 10.1021/bi900085v. Biochemistry. 2009. PMID: 19388699 Free PMC article.
-
Relationship between chaperone activity and oligomeric size of recombinant human alphaA- and alphaB-crystallin: a tryptic digestion study.Proteins. 2004 Nov 15;57(3):610-7. doi: 10.1002/prot.20230. Proteins. 2004. PMID: 15382236
-
Cell biological roles of αB-crystallin.Prog Biophys Mol Biol. 2014 Jul;115(1):3-10. doi: 10.1016/j.pbiomolbio.2014.02.005. Epub 2014 Feb 25. Prog Biophys Mol Biol. 2014. PMID: 24576798 Review.
-
The multifaceted nature of αB-crystallin.Cell Stress Chaperones. 2020 Jul;25(4):639-654. doi: 10.1007/s12192-020-01098-w. Epub 2020 May 7. Cell Stress Chaperones. 2020. PMID: 32383140 Free PMC article. Review.
Cited by
-
Characterization of different-sized human αA-crystallin homomers and implications to Asp151 isomerization.PLoS One. 2024 Jul 11;19(7):e0306856. doi: 10.1371/journal.pone.0306856. eCollection 2024. PLoS One. 2024. PMID: 38991013 Free PMC article.
-
Deletion of Specific Conserved Motifs from the N-Terminal Domain of αB-Crystallin Results in the Activation of Chaperone Functions.Int J Mol Sci. 2022 Jan 20;23(3):1099. doi: 10.3390/ijms23031099. Int J Mol Sci. 2022. PMID: 35163023 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

