Rational engineering of a functional CpG-free ITR for AAV gene therapy
- PMID: 34611321
- PMCID: PMC8983793
- DOI: 10.1038/s41434-021-00296-0
Rational engineering of a functional CpG-free ITR for AAV gene therapy
Abstract
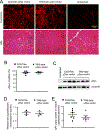
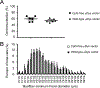
Inverted terminal repeats (ITRs) are the only wild-type components retained in the genome of adeno-associated virus (AAV) vectors. To determine whether ITR modification is a viable approach for AAV vector engineering, we rationally deleted all CpG motifs in the ITR and examined whether CpG elimination compromises AAV-vector production and transduction. Modified ITRs were stable in the plasmid and maintained the CpG-free nature in purified vectors. Replacing the wild-type ITR with the CpG-free ITR did not affect vector genome encapsidation. However, the vector yield was decreased by approximately 3-fold due to reduced vector genome replication. To study the biological potency, we made micro-dystrophin (μDys) AAV vectors carrying either the wild-type ITR or the CpG-free ITR. We delivered the CpG-free μDys vector to one side of the tibialis anterior muscle of dystrophin-null mdx mice and the wild-type μDys vector to the contralateral side. Evaluation at four months after injection showed no difference in the vector genome copy number, microdystrophin expression, and muscle histology and force. Our results suggest that the complete elimination of the CpG motif in the ITR does not affect the biological activity of the AAV vector. CpG-free ITRs could be useful in engineering therapeutic AAV vectors.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures







Similar articles
-
Impact of Inverted Terminal Repeat Integrity on rAAV8 Production Using the Baculovirus/Sf9 Cells System.Hum Gene Ther Methods. 2017 Oct;28(5):277-289. doi: 10.1089/hgtb.2016.133. Hum Gene Ther Methods. 2017. PMID: 28967288 Free PMC article.
-
Adeno-Associated Virus Serotype-Specific Inverted Terminal Repeat Sequence Role in Vector Transgene Expression.Hum Gene Ther. 2020 Feb;31(3-4):151-162. doi: 10.1089/hum.2019.274. Hum Gene Ther. 2020. PMID: 31914802 Free PMC article.
-
Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes.J Virol. 2005 Jan;79(1):364-79. doi: 10.1128/JVI.79.1.364-379.2005. J Virol. 2005. PMID: 15596830 Free PMC article.
-
[Gene therapy for muscular dystrophy].No To Hattatsu. 2004 Mar;36(2):117-23. No To Hattatsu. 2004. PMID: 15031985 Review. Japanese.
-
A User's Guide to the Inverted Terminal Repeats of Adeno-Associated Virus.Hum Gene Ther Methods. 2019 Dec;30(6):206-213. doi: 10.1089/hgtb.2019.276. Hum Gene Ther Methods. 2019. PMID: 31752513 Review.
Cited by
-
Adeno-associated virus-vectored delivery of HIV biologics: the promise of a "single-shot" functional cure for HIV infection.J Virus Erad. 2023 Feb 17;9(1):100316. doi: 10.1016/j.jve.2023.100316. eCollection 2023 Mar. J Virus Erad. 2023. PMID: 36915910 Free PMC article.
-
AAV- based vector improvements unrelated to capsid protein modification.Front Med (Lausanne). 2023 Feb 3;10:1106085. doi: 10.3389/fmed.2023.1106085. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36817775 Free PMC article. Review.
-
Strategies to improve safety profile of AAV vectors.Front Mol Med. 2022 Nov 1;2:1054069. doi: 10.3389/fmmed.2022.1054069. eCollection 2022. Front Mol Med. 2022. PMID: 39086961 Free PMC article. Review.
-
Designing and optimizing AAV-mediated gene therapy for neurodegenerative diseases: from bench to bedside.J Transl Med. 2024 Sep 27;22(1):866. doi: 10.1186/s12967-024-05661-2. J Transl Med. 2024. PMID: 39334366 Free PMC article. Review.
-
Immunogenicity of Recombinant Adeno-Associated Virus (AAV) Vectors for Gene Transfer.BioDrugs. 2023 May;37(3):311-329. doi: 10.1007/s40259-023-00585-7. Epub 2023 Mar 2. BioDrugs. 2023. PMID: 36862289 Free PMC article. Review.
References
-
- Atchison RW, Casto BC, Hammon WM. Adenovirus-associated defective virus particles. Science 1965; 149: 754–6. - PubMed
-
- Carter BJ. Adeno-associated virus and the development of adeno-associated virus vectors: a historical perspective. Mol Ther 2004; 10(6): 981–9. - PubMed
-
- Flotte TR, Berns KI. Adeno-associated virus: a ubiquitous commensal of mammals. Hum Gene Ther 2005; 16(4): 401–7. - PubMed
-
- Muzyczka N Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol 1992; 158: 97–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

