Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus
- PMID: 34611250
- PMCID: PMC8492693
- DOI: 10.1038/s41598-021-99227-7
Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus
Abstract
Dengue poses a global health threat, which will persist without therapeutic intervention. Immunity induced by exposure to one serotype does not confer long-term protection against secondary infection with other serotypes and is potentially capable of enhancing this infection. Although vaccination is believed to induce durable and protective responses against all the dengue virus (DENV) serotypes in order to reduce the burden posed by this virus, the development of a safe and efficacious vaccine remains a challenge. Immunoinformatics and computational vaccinology have been utilized in studies of infectious diseases to provide insight into the host-pathogen interactions thus justifying their use in vaccine development. Since vaccination is the best bet to reduce the burden posed by DENV, this study is aimed at developing a multi-epitope based vaccines for dengue control. Combined approaches of reverse vaccinology and immunoinformatics were utilized to design multi-epitope based vaccine from the sequence of DENV. Specifically, BCPreds and IEDB servers were used to predict the B-cell and T-cell epitopes, respectively. Molecular docking was carried out using Schrödinger, PATCHDOCK and FIREDOCK. Codon optimization and in silico cloning were done using JCAT and SnapGene respectively. Finally, the efficiency and stability of the designed vaccines were assessed by an in silico immune simulation and molecular dynamic simulation, respectively. The predicted epitopes were prioritized using in-house criteria. Four candidate vaccines (DV-1-4) were designed using suitable adjuvant and linkers in addition to the shortlisted epitopes. The binding interactions of these vaccines against the receptors TLR-2, TLR-4, MHC-1 and MHC-2 show that these candidate vaccines perfectly fit into the binding domains of the receptors. In addition, DV-1 has a better binding energies of - 60.07, - 63.40, - 69.89 kcal/mol against MHC-1, TLR-2, and TLR-4, with respect to the other vaccines. All the designed vaccines were highly antigenic, soluble, non-allergenic, non-toxic, flexible, and topologically assessable. The immune simulation analysis showed that DV-1 may elicit specific immune response against dengue virus. Moreover, codon optimization and in silico cloning validated the expressions of all the designed vaccines in E. coli. Finally, the molecular dynamic study shows that DV-1 is stable with minimum RMSF against TLR4. Immunoinformatics tools are now applied to screen genomes of interest for possible vaccine target. The designed vaccine candidates may be further experimentally investigated as potential vaccines capable of providing definitive preventive measure against dengue virus infection.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
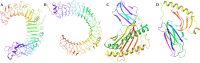
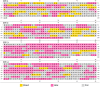

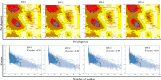



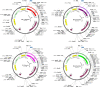

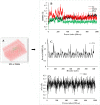
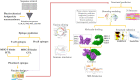
Similar articles
-
Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection.Sci Rep. 2017 Aug 23;7(1):9232. doi: 10.1038/s41598-017-09199-w. Sci Rep. 2017. PMID: 28835708 Free PMC article.
-
Exploring whole proteome to contrive multi-epitope-based vaccine for NeoCoV: An immunoinformtics and in-silico approach.Front Immunol. 2022 Aug 3;13:956776. doi: 10.3389/fimmu.2022.956776. eCollection 2022. Front Immunol. 2022. PMID: 35990651 Free PMC article.
-
Exploring dengue proteome to design an effective epitope-based vaccine against dengue virus.J Biomol Struct Dyn. 2019 Jul;37(10):2546-2563. doi: 10.1080/07391102.2018.1491890. Epub 2018 Nov 17. J Biomol Struct Dyn. 2019. PMID: 30035699
-
Immunogenic multi-epitope-based vaccine development to combat cyclosporiasis of immunocompromised patients applying computational biology method.Exp Parasitol. 2023 May;248:108497. doi: 10.1016/j.exppara.2023.108497. Epub 2023 Mar 10. Exp Parasitol. 2023. PMID: 36906252 Review.
-
Identification and selection of immunodominant B and T cell epitopes for dengue multi-epitope-based vaccine.Med Microbiol Immunol. 2021 Feb;210(1):1-11. doi: 10.1007/s00430-021-00700-x. Epub 2021 Jan 30. Med Microbiol Immunol. 2021. PMID: 33515283 Review.
Cited by
-
Immunoinformatic prediction of the pathogenicity of bovine viral diarrhea virus genotypes: implications for viral virulence determinants, designing novel diagnostic assays and vaccines development.Front Vet Sci. 2023 Jul 6;10:1130147. doi: 10.3389/fvets.2023.1130147. eCollection 2023. Front Vet Sci. 2023. PMID: 37483297 Free PMC article.
-
Annotation of Potential Vaccine Targets and Designing of mRNA-Based Multi-Epitope Vaccine against Lumpy Skin Disease Virus via Reverse Vaccinology and Agent-Based Modeling.Bioengineering (Basel). 2023 Mar 28;10(4):430. doi: 10.3390/bioengineering10040430. Bioengineering (Basel). 2023. PMID: 37106617 Free PMC article.
-
Immunoinformatics analysis of candidate proteins for controlling bovine paratuberculosis.PLoS One. 2022 Nov 21;17(11):e0277751. doi: 10.1371/journal.pone.0277751. eCollection 2022. PLoS One. 2022. PMID: 36409703 Free PMC article.
-
An In Silico Design of a Vaccine against All Serotypes of the Dengue Virus Based on Virtual Screening of B-Cell and T-Cell Epitopes.Biology (Basel). 2024 Aug 30;13(9):681. doi: 10.3390/biology13090681. Biology (Basel). 2024. PMID: 39336108 Free PMC article.
-
Immunoinformatics study to explore dengue (DENV-1) proteome to design multi-epitope vaccine construct by using CD4+ epitopes.J Genet Eng Biotechnol. 2023 Nov 21;21(1):128. doi: 10.1186/s43141-023-00592-9. J Genet Eng Biotechnol. 2023. PMID: 37987878 Free PMC article.
References
-
- Fitzpatrick C, Nwankwo U, Lenk E, de Vlas SJ, Bundy DA. An Investment Case for Ending Neglected Tropical Diseases. The World Bank; 2017. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

