Analysis of the Genetic Diversity Associated With the Drug Resistance and Pathogenicity of Influenza A Virus Isolated in Bangladesh From 2002 to 2019
- PMID: 34603265
- PMCID: PMC8484749
- DOI: 10.3389/fmicb.2021.735305
Analysis of the Genetic Diversity Associated With the Drug Resistance and Pathogenicity of Influenza A Virus Isolated in Bangladesh From 2002 to 2019
Abstract
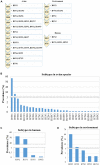
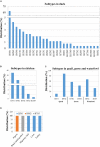
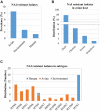
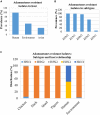
The subtype prevalence, drug resistance- and pathogenicity-associated mutations, and the distribution of the influenza A virus (IAV) isolates identified in Bangladesh from 2002 to 2019 were analyzed using bioinformatic tools. A total of 30 IAV subtypes have been identified in humans (4), avian species (29), and environment (5) in Bangladesh. The predominant subtypes in human and avian species are H1N1/H3N2 and H5N1/H9N2, respectively. However, the subtypes H5N1/H9N2 infecting humans and H3N2/H1N1 infecting avian species have also been identified. Among the avian species, the maximum number of subtypes (27) have been identified in ducks. A 3.56% of the isolates showed neuraminidase inhibitor (NAI) resistance with a prevalence of 8.50, 1.33, and 2.67% in avian species, humans, and the environment, respectively, the following mutations were detected: V116A, I117V, D198N, I223R, S247N, H275Y, and N295S. Prevalence of adamantane-resistant IAVs was 100, 50, and 30.54% in humans, the environment, and avian species, respectively, the subtypes H3N2, H1N1, H9N2, and H5N2 were highly prevalent, with the subtype H5N1 showing a comparatively lower prevalence. Important PB2 mutations such D9N, K526R, A588V, A588I, G590S, Q591R, E627K, K702R, and S714R were identified. A wide range of IAV subtypes have been identified in Bangladesh with a diversified genetic variation in the NA, M2, and PB2 proteins providing drug resistance and enhanced pathogenicity. This study provides a detailed analysis of the subtypes, and the host range of the IAV isolates and the genetic variations related to their proteins, which may aid in the prevention, treatment, and control of IAV infections in Bangladesh, and would serve as a basis for future investigations.
Keywords: Bangladesh; drug resistance; host; influenza A virus; mutations; pathogenicity; subtypes.
Copyright © 2021 Hossain, Akter, Dhole, Saha, Kazi, Majbauddin and Islam.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Effect of influenza H1N1 neuraminidase V116A and I117V mutations on NA activity and sensitivity to NA inhibitors.Antiviral Res. 2019 Sep;169:104539. doi: 10.1016/j.antiviral.2019.104539. Epub 2019 Jun 19. Antiviral Res. 2019. PMID: 31228489
-
New Threats from H7N9 Influenza Virus: Spread and Evolution of High- and Low-Pathogenicity Variants with High Genomic Diversity in Wave Five.J Virol. 2018 May 14;92(11):e00301-18. doi: 10.1128/JVI.00301-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29563296 Free PMC article.
-
Double mutations in the H9N2 avian influenza virus PB2 gene act cooperatively to increase viral host adaptation and replication for human infections.J Gen Virol. 2021 Jun;102(6). doi: 10.1099/jgv.0.001612. J Gen Virol. 2021. PMID: 34061017
-
Drug resistance in influenza A virus: the epidemiology and management.Infect Drug Resist. 2017 Apr 20;10:121-134. doi: 10.2147/IDR.S105473. eCollection 2017. Infect Drug Resist. 2017. PMID: 28458567 Free PMC article. Review.
-
Recent zoonoses caused by influenza A viruses.Rev Sci Tech. 2000 Apr;19(1):197-225. doi: 10.20506/rst.19.1.1220. Rev Sci Tech. 2000. PMID: 11189716 Review.
Cited by
-
PEG-SeNPs as therapeutic agents inhibiting apoptosis and inflammation of cells infected with H1N1 influenza A virus.Sci Rep. 2024 Sep 12;14(1):21318. doi: 10.1038/s41598-024-71486-0. Sci Rep. 2024. PMID: 39266597 Free PMC article.
-
PA-E18G substitution in influenza A virus confers resistance to ZX-7101, a cap-dependent endonuclease inhibitor.Virol Sin. 2023 Aug;38(4):559-567. doi: 10.1016/j.virs.2023.06.002. Epub 2023 Jun 7. Virol Sin. 2023. PMID: 37290559 Free PMC article.
-
A small molecule compound targeting hemagglutinin inhibits influenza A virus and exhibits broad-spectrum antiviral activity.Acta Pharmacol Sin. 2024 Nov;45(11):2380-2393. doi: 10.1038/s41401-024-01331-7. Epub 2024 Jul 10. Acta Pharmacol Sin. 2024. PMID: 38987389
-
Zoonosis and zooanthroponosis of emerging respiratory viruses.Front Cell Infect Microbiol. 2024 Jan 5;13:1232772. doi: 10.3389/fcimb.2023.1232772. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38249300 Free PMC article. Review.
-
Emergence of Eurasian Avian-Like Swine Influenza A (H1N1) virus in a child in Shandong Province, China.BMC Infect Dis. 2024 Jun 1;24(1):550. doi: 10.1186/s12879-024-09441-7. BMC Infect Dis. 2024. PMID: 38824508 Free PMC article.
References
-
- Abed Y., Baz M., Boivin G. (2006). Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir. Ther. 11 971–976. - PubMed
-
- Belongia E. A., Simpson M. D., King J. P., Sundaram M. E., Kelley N. S., Osterholm M. T., et al. (2016). Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect. Dis. 16 942–951. 10.1016/S1473-3099(16)00129-8 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

