TCF4 enhances hepatic metastasis of colorectal cancer by regulating tumor-associated macrophage via CCL2/CCR2 signaling
- PMID: 34580284
- PMCID: PMC8476489
- DOI: 10.1038/s41419-021-04166-w
TCF4 enhances hepatic metastasis of colorectal cancer by regulating tumor-associated macrophage via CCL2/CCR2 signaling
Abstract
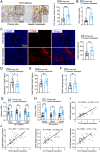
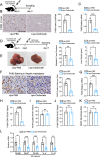
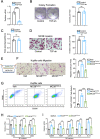
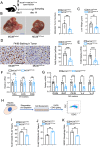
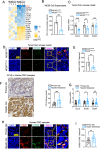
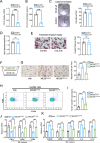
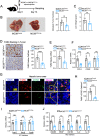
Colorectal cancer (CRC) liver metastasis is a significant clinical problem for which better therapies are urgently needed. Tumor-associated macrophage, a major cell population in the tumor microenvironment, is a known contributor to primary cancer progression and cancer metastasis. Here, we found TAM recruitment and M2 polarization were increased in the hepatic metastatic lesion compared with the primary site of human CRC tissues. Moreover, Pearson correlation analysis showed that TAM recruitment and polarization were closely correlated with the elevated TCF4 expression in the metastatic site. To investigate the role of TCF4 in CRC liver metastasis, we generated a syngeneic mouse model using MC38 cells splenic injection. Results from in vivo experiments and mouse models revealed that TCF4 deficiency in MC38 cells does not affect their proliferation and invasion; however, it reduces TAM infiltration and M2 polarization in the metastasis site. Further studies indicated that these effects are mediated by the TCF4 regulated CCL2 and CCR2 expression. TCF4 or CCL2 silencing in the tumor cells prevent CRC liver metastasis in the mouse model. Altogether, these findings suggest that the TCF4-CCL2-CCR2 axis plays an essential role in CRC liver metastasis by enhancing TAMs recruitment and M2 polarization.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
CCL2-CCR2 axis recruits tumor associated macrophages to induce immune evasion through PD-1 signaling in esophageal carcinogenesis.Mol Cancer. 2020 Feb 27;19(1):41. doi: 10.1186/s12943-020-01165-x. Mol Cancer. 2020. PMID: 32103760 Free PMC article.
-
Dahuang Zhechong Pill suppresses colorectal cancer liver metastasis via ameliorating exosomal CCL2 primed pre-metastatic niche.J Ethnopharmacol. 2019 Jun 28;238:111878. doi: 10.1016/j.jep.2019.111878. Epub 2019 Apr 13. J Ethnopharmacol. 2019. PMID: 30986521
-
NT5DC2 knockdown inhibits colorectal carcinoma progression by repressing metastasis, angiogenesis and tumor-associated macrophage recruitment: A mechanism involving VEGF signaling.Exp Cell Res. 2020 Dec 1;397(1):112311. doi: 10.1016/j.yexcr.2020.112311. Epub 2020 Sep 28. Exp Cell Res. 2020. PMID: 32991874
-
Targeting the CCL2-CCR2 signaling axis in cancer metastasis.Oncotarget. 2016 May 10;7(19):28697-710. doi: 10.18632/oncotarget.7376. Oncotarget. 2016. PMID: 26885690 Free PMC article. Review.
-
Targeting the CCL2/CCR2 Axis in Cancer Immunotherapy: One Stone, Three Birds?Front Immunol. 2021 Nov 3;12:771210. doi: 10.3389/fimmu.2021.771210. eCollection 2021. Front Immunol. 2021. PMID: 34804061 Free PMC article. Review.
Cited by
-
Fn-Dps, a novel virulence factor of Fusobacterium nucleatum, disrupts erythrocytes and promotes metastasis in colorectal cancer.PLoS Pathog. 2023 Jan 24;19(1):e1011096. doi: 10.1371/journal.ppat.1011096. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36693067 Free PMC article.
-
Roles of tumor-associated macrophages in tumor progression: implications on therapeutic strategies.Exp Hematol Oncol. 2021 Dec 29;10(1):60. doi: 10.1186/s40164-021-00252-z. Exp Hematol Oncol. 2021. PMID: 34965886 Free PMC article. Review.
-
Natural compounds modulate the mechanism of action of tumour-associated macrophages against colorectal cancer: a review.J Cancer Res Clin Oncol. 2024 Nov 15;150(11):502. doi: 10.1007/s00432-024-06022-8. J Cancer Res Clin Oncol. 2024. PMID: 39546016 Free PMC article. Review.
-
Research progress of tumor-associated macrophages in immune checkpoint inhibitor tolerance in colorectal cancer.World J Gastrointest Oncol. 2024 Oct 15;16(10):4064-4079. doi: 10.4251/wjgo.v16.i10.4064. World J Gastrointest Oncol. 2024. PMID: 39473964 Free PMC article. Review.
-
Exploiting innate immunity for cancer immunotherapy.Mol Cancer. 2023 Nov 27;22(1):187. doi: 10.1186/s12943-023-01885-w. Mol Cancer. 2023. PMID: 38008741 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

