A Curious Novel Combination of Nucleophosmin (NPM1) Gene Mutations Leading to Aberrant Cytoplasmic Dislocation of NPM1 in Acute Myeloid Leukemia (AML)
- PMID: 34573408
- PMCID: PMC8468273
- DOI: 10.3390/genes12091426
A Curious Novel Combination of Nucleophosmin (NPM1) Gene Mutations Leading to Aberrant Cytoplasmic Dislocation of NPM1 in Acute Myeloid Leukemia (AML)
Abstract
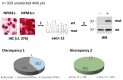
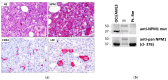
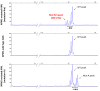
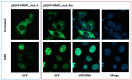
Nucleophosmin (NPM1) mutations occurring in acute myeloid leukemia (AML) (about 50 so far identified) cluster almost exclusively in exon 12 and lead to common changes at the NPM1 mutants C-terminus, i.e., loss of tryptophans 288 and 290 (or 290 alone) and creation of a new nuclear export signal (NES), at the bases of exportin-1(XPO1)-mediated aberrant cytoplasmic NPM1. Immunohistochemistry (IHC) detects cytoplasmic NPM1 and is predictive of the molecular alteration. Besides IHC and molecular sequencing, Western blotting (WB) with anti-NPM1 mutant specific antibodies is another approach to identify NPM1-mutated AML. Here, we show that among 382 AML cases with NPM1 exon 12 mutations, one was not recognized by WB, and describe the discovery of a novel combination of two mutations involving exon 12. This appeared as a conventional mutation A with the known TCTG nucleotides insertion/duplication accompanied by a second event (i.e., an 8-nucleotide deletion occurring 15 nucleotides downstream of the TCTG insertion), resulting in a new C-terminal protein sequence. Strikingly, the sequence included a functional NES ensuring cytoplasmic relocation of the new mutant supporting the role of cytoplasmic NPM1 as critical in AML leukemogenesis.
Keywords: NPM1; acute myeloid leukemia; exportin-1/XPO1; nuclear export signal (NES); nucleophosmin.
Conflict of interest statement
B.F. licensed a patent on
Figures

Similar articles
-
Novel NPM1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in AML.Blood. 2021 Dec 23;138(25):2696-2701. doi: 10.1182/blood.2021012732. Blood. 2021. PMID: 34343258 Free PMC article.
-
A dose-dependent tug of war involving the NPM1 leukaemic mutant, nucleophosmin, and ARF.Leukemia. 2009 Mar;23(3):501-9. doi: 10.1038/leu.2008.326. Epub 2008 Nov 13. Leukemia. 2009. PMID: 19005479
-
Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia.Blood. 2006 Sep 15;108(6):1999-2005. doi: 10.1182/blood-2006-03-007013. Epub 2006 May 23. Blood. 2006. PMID: 16720834
-
Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications.Leukemia. 2009 Oct;23(10):1731-43. doi: 10.1038/leu.2009.124. Epub 2009 Jun 11. Leukemia. 2009. PMID: 19516275 Review.
-
Nucleophosmin 1: from its pathogenic role to a tantalizing therapeutic target in acute myeloid leukemia.Hematology. 2022 Dec;27(1):609-619. doi: 10.1080/16078454.2022.2067939. Hematology. 2022. PMID: 35621728 Review.
Cited by
-
Nucleophosmin promotes lung adenocarcinoma cell proliferation, migration and invasion by activating the EGFR/MAPK signaling pathway.Oncol Rep. 2023 Jun;49(6):126. doi: 10.3892/or.2023.8563. Epub 2023 May 11. Oncol Rep. 2023. PMID: 37165929 Free PMC article.
References
-
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

