Reproducible Lipid Alterations in Patient-Derived Breast Cancer Xenograft FFPE Tissue Identified with MALDI MSI for Pre-Clinical and Clinical Application
- PMID: 34564393
- PMCID: PMC8467053
- DOI: 10.3390/metabo11090577
Reproducible Lipid Alterations in Patient-Derived Breast Cancer Xenograft FFPE Tissue Identified with MALDI MSI for Pre-Clinical and Clinical Application
Abstract
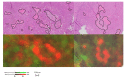
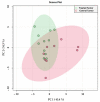
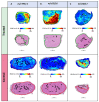
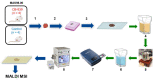
The association between lipid metabolism and long-term outcomes is relevant for tumor diagnosis and therapy. Archival material such as formalin-fixed and paraffin embedded (FFPE) tissues is a highly valuable resource for this aim as it is linked to long-term clinical follow-up. Therefore, there is a need to develop robust methodologies able to detect lipids in FFPE material and correlate them with clinical outcomes. In this work, lipidic alterations were investigated in patient-derived xenograft of breast cancer by using a matrix-assisted laser desorption ionization mass spectrometry (MALDI MSI) based workflow that included antigen retrieval as a sample preparation step. We evaluated technical reproducibility, spatial metabolic differentiation within tissue compartments, and treatment response induced by a glutaminase inhibitor (CB-839). This protocol shows a good inter-day robustness (CV = 26 ± 12%). Several lipids could reliably distinguish necrotic and tumor regions across the technical replicates. Moreover, this protocol identified distinct alterations in the tissue lipidome of xenograft treated with glutaminase inhibitors. In conclusion, lipidic alterations in FFPE tissue of breast cancer xenograft observed in this study are a step-forward to a robust and reproducible MALDI-MSI based workflow for pre-clinical and clinical applications.
Keywords: FFPE tissue; MALDI MSI; breast cancer; diagnosis; lipidomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Visualization of Small Intact Proteins in Breast Cancer FFPE Tissue.Methods Mol Biol. 2023;2688:161-172. doi: 10.1007/978-1-0716-3319-9_14. Methods Mol Biol. 2023. PMID: 37410292
-
Antigen Retrieval and Its Effect on the MALDI-MSI of Lipids in Formalin-Fixed Paraffin-Embedded Tissue.J Am Soc Mass Spectrom. 2020 Aug 5;31(8):1619-1624. doi: 10.1021/jasms.0c00208. Epub 2020 Jul 22. J Am Soc Mass Spectrom. 2020. PMID: 32678590 Free PMC article.
-
Sample preparation of formalin-fixed paraffin-embedded tissue sections for MALDI-mass spectrometry imaging.Anal Bioanal Chem. 2020 Feb;412(6):1263-1275. doi: 10.1007/s00216-019-02296-x. Epub 2020 Jan 28. Anal Bioanal Chem. 2020. PMID: 31989198 Free PMC article.
-
[Mass spectrometry imaging technology and its application in breast cancer research].Se Pu. 2021 Jun;39(6):578-587. doi: 10.3724/SP.J.1123.2020.10005. Se Pu. 2021. PMID: 34227318 Free PMC article. Review. Chinese.
-
MALDI IMS and Cancer Tissue Microarrays.Adv Cancer Res. 2017;134:173-200. doi: 10.1016/bs.acr.2016.11.007. Epub 2017 Jan 12. Adv Cancer Res. 2017. PMID: 28110650 Review.
Cited by
-
The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer.Int J Mol Sci. 2024 Sep 29;25(19):10503. doi: 10.3390/ijms251910503. Int J Mol Sci. 2024. PMID: 39408832 Free PMC article. Review.
-
Sample Preparation for Metabolite Detection in Mass Spectrometry Imaging.Methods Mol Biol. 2023;2688:135-146. doi: 10.1007/978-1-0716-3319-9_12. Methods Mol Biol. 2023. PMID: 37410290
-
Application of spatial-omics to the classification of kidney biopsy samples in transplantation.Nat Rev Nephrol. 2024 Nov;20(11):755-766. doi: 10.1038/s41581-024-00861-x. Epub 2024 Jul 4. Nat Rev Nephrol. 2024. PMID: 38965417 Review.
-
Visualization of Small Intact Proteins in Breast Cancer FFPE Tissue.Methods Mol Biol. 2023;2688:161-172. doi: 10.1007/978-1-0716-3319-9_14. Methods Mol Biol. 2023. PMID: 37410292
-
rMSIfragment: improving MALDI-MSI lipidomics through automated in-source fragment annotation.J Cheminform. 2023 Sep 15;15(1):80. doi: 10.1186/s13321-023-00756-2. J Cheminform. 2023. PMID: 37715285 Free PMC article.
References
-
- Graça G., Lau C.-H.E., Gonçalves L.G. Exploring Cancer Metabolism: Applications of Metabolomics and Metabolic Phenotyping in Cancer Research and Diagnostics. In: Serpa J., editor. Tumor Microenvironment: The Main Driver of Metabolic Adaptation. Springer International Publishing; Cham, Switzerland: 2020. pp. 367–385. - PubMed
-
- Dey P., Kimmelman A.C., DePinho R.A. Metabolic Codependencies in the Tumor Microenvironment. Cancer Discov. 2021;11:1067–1081. doi: 10.1158/2159-8290.CD-20-1211. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

