Structure of a Ty1 restriction factor reveals the molecular basis of transposition copy number control
- PMID: 34552077
- PMCID: PMC8458377
- DOI: 10.1038/s41467-021-25849-0
Structure of a Ty1 restriction factor reveals the molecular basis of transposition copy number control
Abstract
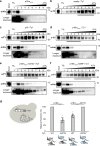
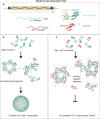
Excessive replication of Saccharomyces cerevisiae Ty1 retrotransposons is regulated by Copy Number Control, a process requiring the p22/p18 protein produced from a sub-genomic transcript initiated within Ty1 GAG. In retrotransposition, Gag performs the capsid functions required for replication and re-integration. To minimize genomic damage, p22/p18 interrupts virus-like particle function by interaction with Gag. Here, we present structural, biophysical and genetic analyses of p18m, a minimal fragment of Gag that restricts transposition. The 2.8 Å crystal structure of p18m reveals an all α-helical protein related to mammalian and insect ARC proteins. p18m retains the capacity to dimerise in solution and the crystal structures reveal two exclusive dimer interfaces. We probe our findings through biophysical analysis of interface mutants as well as Ty1 transposition and p18m restriction in vivo. Our data provide insight into Ty1 Gag structure and suggest how p22/p18 might function in restriction through a blocking-of-assembly mechanism.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing financial or non-financial interests.
Figures

Similar articles
-
A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control.J Virol. 2015 Apr;89(7):3922-38. doi: 10.1128/JVI.03060-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609815 Free PMC article.
-
The Ty1 Retrotransposon Restriction Factor p22 Targets Gag.PLoS Genet. 2015 Oct 9;11(10):e1005571. doi: 10.1371/journal.pgen.1005571. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26451601 Free PMC article.
-
Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae.Genetics. 2017 Dec;207(4):1441-1456. doi: 10.1534/genetics.117.300388. Epub 2017 Oct 18. Genetics. 2017. PMID: 29046400 Free PMC article.
-
A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces.Curr Genet. 2016 May;62(2):321-9. doi: 10.1007/s00294-015-0550-6. Epub 2015 Dec 9. Curr Genet. 2016. PMID: 26650614 Free PMC article. Review.
-
Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element.Cytogenet Genome Res. 2005;110(1-4):269-87. doi: 10.1159/000084960. Cytogenet Genome Res. 2005. PMID: 16093680 Review.
Cited by
-
An interchangeable prion-like domain is required for Ty1 retrotransposition.Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2303358120. doi: 10.1073/pnas.2303358120. Epub 2023 Jul 17. Proc Natl Acad Sci U S A. 2023. PMID: 37459521 Free PMC article.
-
A high-quality reference genome for the fission yeast Schizosaccharomyces osmophilus.G3 (Bethesda). 2023 Apr 11;13(4):jkad028. doi: 10.1093/g3journal/jkad028. G3 (Bethesda). 2023. PMID: 36748990 Free PMC article.
-
Evolution of a Restriction Factor by Domestication of a Yeast Retrotransposon.Mol Biol Evol. 2024 Mar 1;41(3):msae050. doi: 10.1093/molbev/msae050. Mol Biol Evol. 2024. PMID: 38442736 Free PMC article.
-
An interchangeable prion-like domain is required for Ty1 retrotransposition.bioRxiv [Preprint]. 2023 Feb 27:2023.02.27.530227. doi: 10.1101/2023.02.27.530227. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2303358120. doi: 10.1073/pnas.2303358120 PMID: 36909481 Free PMC article. Updated. Preprint.
-
PTGS is dispensable for the initiation of epigenetic silencing of an active transposon in Arabidopsis.EMBO Rep. 2024 Dec;25(12):5780-5809. doi: 10.1038/s44319-024-00304-5. Epub 2024 Nov 7. EMBO Rep. 2024. PMID: 39511423 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

