Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer
- PMID: 34549974
- PMCID: PMC8610584
- DOI: 10.1128/JVI.00529-21
Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer
Abstract
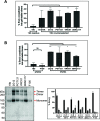
The functional human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer [(gp120/gp41)3] is produced by cleavage of a conformationally flexible gp160 precursor. gp160 cleavage or the binding of BMS-806, an entry inhibitor, stabilizes the pretriggered, "closed" (state 1) conformation recognized by rarely elicited broadly neutralizing antibodies. Poorly neutralizing antibodies (pNAbs) elicited at high titers during natural infection recognize more "open" Env conformations (states 2 and 3) induced by binding the receptor, CD4. We found that BMS-806 treatment and cross-linking decreased the exposure of pNAb epitopes on cell surface gp160; however, after detergent solubilization, cross-linked and BMS-806-treated gp160 sampled non-state-1 conformations that could be recognized by pNAbs. Cryo-electron microscopy of the purified BMS-806-bound gp160 revealed two hitherto unknown asymmetric trimer conformations, providing insights into the allosteric coupling between trimer opening and structural variation in the gp41 HR1N region. The individual protomer structures in the asymmetric gp160 trimers resemble those of other genetically modified or antibody-bound cleaved HIV-1 Env trimers, which have been suggested to assume state-2-like conformations. Asymmetry of the uncleaved Env potentially exposes surfaces of the trimer to pNAbs. To evaluate the effect of stabilizing a state-1-like conformation of the membrane Env precursor, we treated cells expressing wild-type HIV-1 Env with BMS-806. BMS-806 treatment decreased both gp160 cleavage and the addition of complex glycans, implying that gp160 conformational flexibility contributes to the efficiency of these processes. Selective pressure to maintain flexibility in the precursor of functional Env allows the uncleaved Env to sample asymmetric conformations that potentially skew host antibody responses toward pNAbs. IMPORTANCE The envelope glycoprotein (Env) trimers on the surface of human immunodeficiency virus (HIV-1) mediate the entry of the virus into host cells and serve as targets for neutralizing antibodies. The functional Env trimer is produced by cleavage of the gp160 precursor in the infected cell. We found that the HIV-1 Env precursor is highly plastic, allowing it to assume different asymmetric shapes. This conformational plasticity is potentially important for Env cleavage and proper modification by sugars. Having a flexible, asymmetric Env precursor that can misdirect host antibody responses without compromising virus infectivity would be an advantage for a persistent virus like HIV-1.
Keywords: Env; antibody; asymmetry; cleavage; conformation; cryo-electron microscopy; furin; processing; structure.
Figures


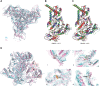

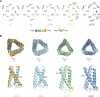




Similar articles
-
Inducible cell lines producing replication-defective human immunodeficiency virus particles containing envelope glycoproteins stabilized in a pretriggered conformation.J Virol. 2024 Dec 17;98(12):e0172024. doi: 10.1128/jvi.01720-24. Epub 2024 Nov 7. J Virol. 2024. PMID: 39508605
-
Comparison of Uncleaved and Mature Human Immunodeficiency Virus Membrane Envelope Glycoprotein Trimers.J Virol. 2018 May 29;92(12):e00277-18. doi: 10.1128/JVI.00277-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618643 Free PMC article.
-
Dual Pathways of Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Trafficking Modulate the Selective Exclusion of Uncleaved Oligomers from Virions.J Virol. 2021 Jan 13;95(3):e01369-20. doi: 10.1128/JVI.01369-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33148792 Free PMC article.
-
Conformation-Dependent Interactions Between HIV-1 Envelope Glycoproteins and Broadly Neutralizing Antibodies.AIDS Res Hum Retroviruses. 2018 Sep;34(9):794-803. doi: 10.1089/AID.2018.0102. Epub 2018 Jul 17. AIDS Res Hum Retroviruses. 2018. PMID: 29905080 Review.
-
Structure-guided envelope trimer design in HIV-1 vaccine development: a narrative review.J Int AIDS Soc. 2021 Nov;24 Suppl 7(Suppl 7):e25797. doi: 10.1002/jia2.25797. J Int AIDS Soc. 2021. PMID: 34806305 Free PMC article. Review.
Cited by
-
Conformations of membrane human immunodeficiency virus (HIV-1) envelope glycoproteins solubilized in Amphipol A18 lipid-nanodiscs.J Virol. 2024 Oct 22;98(10):e0063124. doi: 10.1128/jvi.00631-24. Epub 2024 Sep 9. J Virol. 2024. PMID: 39248459
-
Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.Nat Commun. 2023 Oct 23;14(1):6710. doi: 10.1038/s41467-023-42500-2. Nat Commun. 2023. PMID: 37872202 Free PMC article.
-
HIV-1 Envelope Glycoproteins Proteolytic Cleavage Protects Infected Cells from ADCC Mediated by Plasma from Infected Individuals.Viruses. 2021 Nov 6;13(11):2236. doi: 10.3390/v13112236. Viruses. 2021. PMID: 34835042 Free PMC article.
-
Stoichiometry of HIV-1 Envelope Glycoprotein Protomers with Changes That Stabilize or Destabilize the Pretriggered Conformation.bioRxiv [Preprint]. 2024 Oct 25:2024.10.25.620268. doi: 10.1101/2024.10.25.620268. bioRxiv. 2024. PMID: 39484577 Free PMC article. Preprint.
-
Alterations in gp120 glycans or the gp41 fusion peptide-proximal region modulate the stability of the human immunodeficiency virus (HIV-1) envelope glycoprotein pretriggered conformation.J Virol. 2023 Sep 28;97(9):e0059223. doi: 10.1128/jvi.00592-23. Epub 2023 Sep 11. J Virol. 2023. PMID: 37696048 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- UM1 AI100645/AI/NIAID NIH HHS/United States
- P01 AI150471/AI/NIAID NIH HHS/United States
- AI027767/HHS | National Institutes of Health (NIH)
- R01 AI125093/AI/NIAID NIH HHS/United States
- P01 GM056550/GM/NIGMS NIH HHS/United States
- AI125093/HHS | National Institutes of Health (NIH)
- 109998-67-RKVA/amfAR, The Foundation for AIDS Research (amfAR)
- R01 AI145547/AI/NIAID NIH HHS/United States
- R01 AI093256/AI/NIAID NIH HHS/United States
- AI093256/HHS | National Institutes of Health (NIH)
- 11774012/National Natural Science Foundation of China (NSFC)
- P30 AI027767/AI/NIAID NIH HHS/United States
- R01 AI124982/AI/NIAID NIH HHS/United States
- Z180016/Z18J008/Natural Science Foundation of Beijing Municipality
LinkOut - more resources
Full Text Sources
Research Materials

