Ancient viral integrations in marsupials: a potential antiviral defence
- PMID: 34548931
- PMCID: PMC8449507
- DOI: 10.1093/ve/veab076
Ancient viral integrations in marsupials: a potential antiviral defence
Abstract
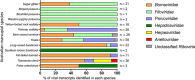
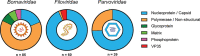
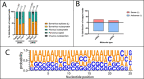
Marsupial viruses are understudied compared to their eutherian mammal counterparts, although they may pose severe threats to vulnerable marsupial populations. Genomic viral integrations, termed 'endogenous viral elements' (EVEs), could protect the host from infection. It is widely known past viral infections and EVEs play an active role in antiviral defence in invertebrates and plants. This study aimed to characterise actively transcribed EVEs in Australian marsupial species, because they may play an integral role in cellular defence against viruses. This study screened publicly available RNA sequencing data sets (n = 35) and characterised 200 viral transcripts from thirteen Australian marsupial species. Of the 200 transcripts, 188 originated from either Bornaviridae, Filoviridae, or Parvoviridae EVEs. The other twelve transcripts were from putative active infections from members of the Herpesviridae and Anelloviridae, and Hepadnaviridae. EVE transcripts (n = 188) were mapped to marsupial genomes (where available, n = 5/13) to identify the genomic insertion sites. Of the 188 transcripts, 117 mapped to 39 EVEs within the koala, bare-nosed wombat, tammar wallaby, brushtail possum, and Tasmanian devil genomes. The remaining eight animals had no available genome (transcripts n = 71). Every marsupial has Bornaviridae, Filoviridae, and Parvoviridae EVEs, a trend widely observed in eutherian mammals. Whilst eutherian bornavirus EVEs are predominantly nucleoprotein-derived, marsupial bornavirus EVEs demonstrate a surprising replicase gene bias. We predicted these widely distributed EVEs were conserved within marsupials from ancient germline integrations, as many were over 65 million years old. One bornavirus replicase EVE, present in six marsupial genomes, was estimated to be 160 million years old, predating the American-Australian marsupial split. We considered transcription of these EVEs through small non-coding RNA as an ancient viral defence. Consistent with this, in koala small RNA sequence data sets, we detected Bornaviridae replicase and Filoviridae nucleoprotein produced small RNA. These were enriched in testis tissue, suggesting they could protect marsupials from vertically transmitted viral integrations.
Keywords: RNAi; endogenous viral element; marsupial; paleovirology; piRNA; small RNA; virus evolution.
© The Author(s) 2021. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Endogenous Viral Elements Are Widespread in Arthropod Genomes and Commonly Give Rise to PIWI-Interacting RNAs.J Virol. 2019 Mar 5;93(6):e02124-18. doi: 10.1128/JVI.02124-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567990 Free PMC article.
-
Paleovirology of bornaviruses: What can be learned from molecular fossils of bornaviruses.Virus Res. 2019 Mar;262:2-9. doi: 10.1016/j.virusres.2018.04.006. Epub 2018 Apr 6. Virus Res. 2019. PMID: 29630909 Review.
-
A Human Endogenous Bornavirus-Like Nucleoprotein Encodes a Mitochondrial Protein Associated with Cell Viability.J Virol. 2021 Jun 24;95(14):e0203020. doi: 10.1128/JVI.02030-20. Epub 2021 Jun 24. J Virol. 2021. PMID: 33952640 Free PMC article.
-
Evolutionary Analysis of Placental Orthologues Reveals Two Ancient DNA Virus Integrations.J Virol. 2022 Nov 23;96(22):e0093322. doi: 10.1128/jvi.00933-22. Epub 2022 Oct 27. J Virol. 2022. PMID: 36300941 Free PMC article.
-
Viral Sequences Are Repurposed for Controlling Antiviral Responses as Non-Retroviral Endogenous Viral Elements.Acta Med Okayama. 2022 Oct;76(5):503-510. doi: 10.18926/AMO/64025. Acta Med Okayama. 2022. PMID: 36352796 Review.
Cited by
-
Deep mining reveals the diversity of endogenous viral elements in vertebrate genomes.Nat Microbiol. 2024 Nov;9(11):3013-3024. doi: 10.1038/s41564-024-01825-4. Epub 2024 Oct 22. Nat Microbiol. 2024. PMID: 39438719 Free PMC article.
-
Novel viruses discovered in the transcriptomes of agnathan fish.J Fish Dis. 2022 Jun;45(6):931-938. doi: 10.1111/jfd.13602. Epub 2022 Mar 2. J Fish Dis. 2022. PMID: 35235679 Free PMC article. No abstract available.
-
Identification of a novel polyomavirus from a marsupial host.Virus Evol. 2022 Oct 6;8(2):veac096. doi: 10.1093/ve/veac096. eCollection 2022. Virus Evol. 2022. PMID: 36381233 Free PMC article.
-
Genomic transfers help to decipher the ancient evolution of filoviruses and interactions with vertebrate hosts.PLoS Pathog. 2024 Sep 3;20(9):e1011864. doi: 10.1371/journal.ppat.1011864. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39226335 Free PMC article.
-
Divergent hepaciviruses, delta-like viruses, and a chu-like virus in Australian marsupial carnivores (dasyurids).Virus Evol. 2023 Oct 14;9(2):vead061. doi: 10.1093/ve/vead061. eCollection 2023. Virus Evol. 2023. PMID: 37941997 Free PMC article.
References
-
- Amery-Gale J. et al. (2014) ‘Detection and Identification of a Gammaherpesvirus in Antechinus spp. In Australia’, Journal of Wildlife Diseases, 50: 334–9. - PubMed
-
- Andrews S. (2010) ‘FastQC: A Quality Control Tool for High Throughput Sequence Data’, Babraham Bioinformatics. <https://www.bioinformatics.babraham.ac.uk/projects/fastqc/> accessed 30 Aug 2021.
LinkOut - more resources
Full Text Sources
Miscellaneous

