Transcriptome-wide Dynamics of m6A mRNA Methylation During Porcine Spermatogenesis
- PMID: 34543723
- PMCID: PMC10787014
- DOI: 10.1016/j.gpb.2021.08.006
Transcriptome-wide Dynamics of m6A mRNA Methylation During Porcine Spermatogenesis
Abstract
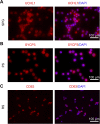
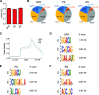
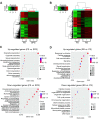
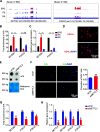
Spermatogenesis is a continual process that occurs in the testes, in which diploid spermatogonial stem cells (SSCs) differentiate and generate haploid spermatozoa. This highly efficient and intricate process is orchestrated at multiple levels. N6-methyladenosine (m6A), an epigenetic modification prevalent in mRNAs, is implicated in the transcriptional regulation during spermatogenesis. However, the dynamics of m6A modification in non-rodent mammalian species remains unclear. Here, we systematically investigated the profile and role of m6A during spermatogenesis in pigs. By analyzing the transcriptomic distribution of m6A in spermatogonia, spermatocytes, and round spermatids, we identified a globally conserved m6A pattern between porcine and murine genes with spermatogenic function. We found that m6A was enriched in a group of genes that specifically encode the metabolic enzymes and regulators. In addition, transcriptomes in porcine male germ cells could be subjected to the m6A modification. Our data show that m6A plays the regulatory roles during spermatogenesis in pigs, which is similar to that in mice. Illustrations of this point are three genes (SETDB1, FOXO1, and FOXO3) that are crucial to the determination of the fate of SSCs. To the best of our knowledge, this study for the first time uncovers the expression profile and role of m6A during spermatogenesis in large animals and provides insights into the intricate transcriptional regulation underlying the lifelong male fertility in non-rodent mammalian species.
Keywords: N(6)-methyladenosine; Pig; SETDB1; Spermatogenesis; Spermatogonial stem cell.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have declared no competing interests.
Figures


Similar articles
-
Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis.Cell Res. 2017 Oct;27(10):1216-1230. doi: 10.1038/cr.2017.117. Epub 2017 Sep 15. Cell Res. 2017. PMID: 28914256 Free PMC article.
-
[The Roles of N6-Methyladenosine Modification and Its Regulators in Male Reproduction].Sichuan Da Xue Xue Bao Yi Xue Ban. 2024 May 20;55(3):527-534. doi: 10.12182/20240560103. Sichuan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38948273 Free PMC article. Review. Chinese.
-
Single-Nucleus RNA-Seq Reveals Spermatogonial Stem Cell Developmental Pattern in Shaziling Pigs.Biomolecules. 2024 May 21;14(6):607. doi: 10.3390/biom14060607. Biomolecules. 2024. PMID: 38927011 Free PMC article.
-
Single-cell transcriptome analyses reveal critical regulators of spermatogonial stem cell fate transitions.BMC Genomics. 2024 Feb 3;25(1):138. doi: 10.1186/s12864-024-10072-0. BMC Genomics. 2024. PMID: 38310206 Free PMC article.
-
Mechanisms regulating mammalian spermatogenesis and fertility recovery following germ cell depletion.Cell Mol Life Sci. 2019 Oct;76(20):4071-4102. doi: 10.1007/s00018-019-03201-6. Epub 2019 Jun 28. Cell Mol Life Sci. 2019. PMID: 31254043 Free PMC article. Review.
Cited by
-
SETDB1 Regulates Porcine Spermatogonial Adhesion and Proliferation through Modulating MMP3/10 Transcription.Cells. 2022 Jan 22;11(3):370. doi: 10.3390/cells11030370. Cells. 2022. PMID: 35159180 Free PMC article.
-
Transcriptome Studies Reveal the N6-Methyladenosine Differences in Testis of Yaks at Juvenile and Sexual Maturity Stages.Animals (Basel). 2023 Sep 5;13(18):2815. doi: 10.3390/ani13182815. Animals (Basel). 2023. PMID: 37760215 Free PMC article.
-
Emerging role of m6A methylation modification in ovarian cancer.Cancer Cell Int. 2021 Dec 11;21(1):663. doi: 10.1186/s12935-021-02371-3. Cancer Cell Int. 2021. PMID: 34895230 Free PMC article. Review.
-
Characterization of sexual maturity-associated N6-methyladenosine in boar testes.BMC Genomics. 2024 May 7;25(1):447. doi: 10.1186/s12864-024-10343-w. BMC Genomics. 2024. PMID: 38714941 Free PMC article.
-
Research Progress on the Role of M6A in Regulating Economic Traits in Livestock.Int J Mol Sci. 2024 Jul 31;25(15):8365. doi: 10.3390/ijms25158365. Int J Mol Sci. 2024. PMID: 39125935 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

