CombiFlow: combinatorial AML-specific plasma membrane expression profiles allow longitudinal tracking of clones
- PMID: 34543390
- PMCID: PMC9006304
- DOI: 10.1182/bloodadvances.2021005018
CombiFlow: combinatorial AML-specific plasma membrane expression profiles allow longitudinal tracking of clones
Abstract
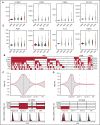
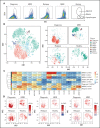
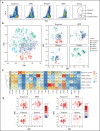
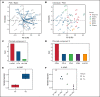
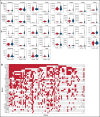
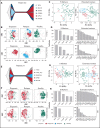
Acute myeloid leukemia (AML) often presents as an oligoclonal disease whereby multiple genetically distinct subclones can coexist within patients. Differences in signaling and drug sensitivity of such subclones complicate treatment and warrant tools to identify them and track disease progression. We previously identified >50 AML-specific plasma membrane (PM) proteins, and 7 of these (CD82, CD97, FLT3, IL1RAP, TIM3, CD25, and CD123) were implemented in routine diagnostics in patients with AML (n = 256) and myelodysplastic syndrome (n = 33). We developed a pipeline termed CombiFlow in which expression data of multiple PM markers is merged, allowing a principal component-based analysis to identify distinctive marker expression profiles and to generate single-cell t-distributed stochastic neighbor embedding landscapes to longitudinally track clonal evolution. Positivity for one or more of the markers after 2 courses of intensive chemotherapy predicted a shorter relapse-free survival, supporting a role for these markers in measurable residual disease (MRD) detection. CombiFlow also allowed the tracking of clonal evolution in paired diagnosis and relapse samples. Extending the panel to 36 AML-specific markers further refined the CombiFlow pipeline. In conclusion, CombiFlow provides a valuable tool in the diagnosis, MRD detection, clonal tracking, and understanding of clonal heterogeneity in AML.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures



Similar articles
-
CombiFlow: Flow cytometry-based identification and characterization of genetically and functionally distinct AML subclones.STAR Protoc. 2021 Sep 27;2(4):100864. doi: 10.1016/j.xpro.2021.100864. eCollection 2021 Dec 17. STAR Protoc. 2021. PMID: 34622222 Free PMC article.
-
Single-cell mutational profiling enhances the clinical evaluation of AML MRD.Blood Adv. 2020 Mar 10;4(5):943-952. doi: 10.1182/bloodadvances.2019001181. Blood Adv. 2020. PMID: 32150611 Free PMC article.
-
Prospective Isolation and Characterization of Genetically and Functionally Distinct AML Subclones.Cancer Cell. 2018 Oct 8;34(4):674-689.e8. doi: 10.1016/j.ccell.2018.08.014. Epub 2018 Sep 20. Cancer Cell. 2018. PMID: 30245083
-
Clonal evolution of acute myeloid leukemia from diagnosis to relapse.Genes Chromosomes Cancer. 2019 Dec;58(12):839-849. doi: 10.1002/gcc.22806. Epub 2019 Sep 3. Genes Chromosomes Cancer. 2019. PMID: 31478278 Free PMC article. Review.
-
Minimal/Measurable Residual Disease Monitoring in NPM1-Mutated Acute Myeloid Leukemia: A Clinical Viewpoint and Perspectives.Int J Mol Sci. 2018 Nov 6;19(11):3492. doi: 10.3390/ijms19113492. Int J Mol Sci. 2018. PMID: 30404199 Free PMC article. Review.
Cited by
-
To Detach, Migrate, Adhere, and Metastasize: CD97/ADGRE5 in Cancer.Cells. 2022 May 4;11(9):1538. doi: 10.3390/cells11091538. Cells. 2022. PMID: 35563846 Free PMC article. Review.
-
CD123 a Therapeutic Target for Acute Myeloid Leukemia and Blastic Plasmocytoid Dendritic Neoplasm.Int J Mol Sci. 2023 Feb 1;24(3):2718. doi: 10.3390/ijms24032718. Int J Mol Sci. 2023. PMID: 36769040 Free PMC article. Review.
-
Anti-IL-1RAP scFv-mSA-S19-TAT fusion carrier as a multifunctional platform for versatile delivery of biotinylated payloads to myeloid leukemia cells.Sci Rep. 2024 Oct 23;14(1):25080. doi: 10.1038/s41598-024-76851-7. Sci Rep. 2024. PMID: 39443595 Free PMC article.
-
Chimeric antigen receptor T-cells targeting IL-1RAP: a promising new cellular immunotherapy to treat acute myeloid leukemia.J Immunother Cancer. 2022 Jul;10(7):e004222. doi: 10.1136/jitc-2021-004222. J Immunother Cancer. 2022. PMID: 35803613 Free PMC article.
References
-
- de Lima M, Porter DL, Battiwalla M, et al. . Proceedings from the National Cancer Institute's Second International Workshop on the Biology, Prevention, and Treatment of Relapse After Hematopoietic Stem Cell Transplantation: part III. Prevention and treatment of relapse after allogeneic transplantation. Biol Blood Marrow Transplant. 2014;20(1):4-13. - PMC - PubMed
-
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997; 3(7):730-737. - PubMed
-
- Valent P, Bonnet D, De Maria R, et al. . Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12(11):767-775. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

