Evolutionary dynamics of circular RNAs in primates
- PMID: 34542404
- PMCID: PMC8516421
- DOI: 10.7554/eLife.69148
Evolutionary dynamics of circular RNAs in primates
Abstract
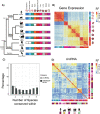
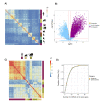
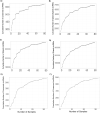
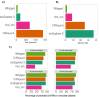
Many primate genes produce circular RNAs (circRNAs). However, the extent of circRNA conservation between closely related species remains unclear. By comparing tissue-specific transcriptomes across over 70 million years of primate evolution, we identify that within 3 million years circRNA expression profiles diverged such that they are more related to species identity than organ type. However, our analysis also revealed a subset of circRNAs with conserved neural expression across tens of millions of years of evolution. By comparing to species-specific circRNAs, we identified that the downstream intron of the conserved circRNAs display a dramatic lengthening during evolution due to the insertion of novel retrotransposons. Our work provides comparative analyses of the mechanisms promoting circRNAs to generate increased transcriptomic complexity in primates.
Keywords: RNA; circRNA; comparative genomics; evolution; evolutionary biology; genetics; genomics; human; non-coding RNAs; rhesus macaque.
© 2021, Santos-Rodriguez et al.
Conflict of interest statement
GS, IV, RW No competing interests declared
Figures

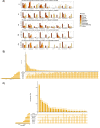

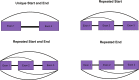
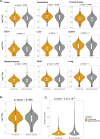

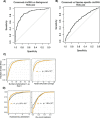
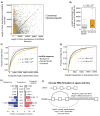
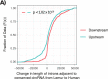
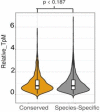
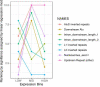
Similar articles
-
Circular RNA repertoires are associated with evolutionarily young transposable elements.Elife. 2021 Sep 20;10:e67991. doi: 10.7554/eLife.67991. Elife. 2021. PMID: 34542406 Free PMC article.
-
Neural circular transcriptomes across mammalian species.Genomics. 2020 Mar;112(2):1162-1166. doi: 10.1016/j.ygeno.2019.06.030. Epub 2019 Jun 28. Genomics. 2020. PMID: 31255695 Review.
-
Analysis of pig transcriptomes suggests a global regulation mechanism enabling temporary bursts of circular RNAs.RNA Biol. 2019 Sep;16(9):1190-1204. doi: 10.1080/15476286.2019.1621621. Epub 2019 Jun 3. RNA Biol. 2019. PMID: 31120323 Free PMC article.
-
Comparative Analysis of Gammaherpesvirus Circular RNA Repertoires: Conserved and Unique Viral Circular RNAs.J Virol. 2019 Mar 5;93(6):e01952-18. doi: 10.1128/JVI.01952-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567979 Free PMC article.
-
Circular RNAs in cell differentiation and development.Development. 2020 Aug 24;147(16):dev182725. doi: 10.1242/dev.182725. Development. 2020. PMID: 32839270 Review.
Cited by
-
Circular RNA repertoires are associated with evolutionarily young transposable elements.Elife. 2021 Sep 20;10:e67991. doi: 10.7554/eLife.67991. Elife. 2021. PMID: 34542406 Free PMC article.
-
The roles of circular RNAs in nerve injury and repair.Front Mol Neurosci. 2024 Jul 15;17:1419520. doi: 10.3389/fnmol.2024.1419520. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39077756 Free PMC article. Review.
-
Human brain evolution: Emerging roles for regulatory DNA and RNA.Curr Opin Neurobiol. 2021 Dec;71:170-177. doi: 10.1016/j.conb.2021.11.005. Epub 2021 Nov 30. Curr Opin Neurobiol. 2021. PMID: 34861533 Free PMC article. Review.
-
Circular RNA expression in turkey skeletal muscle satellite cells is significantly altered by thermal challenge.Front Physiol. 2024 Sep 18;15:1476487. doi: 10.3389/fphys.2024.1476487. eCollection 2024. Front Physiol. 2024. PMID: 39359572 Free PMC article.
-
Circular RNA in cancer.Nat Rev Cancer. 2024 Sep;24(9):597-613. doi: 10.1038/s41568-024-00721-7. Epub 2024 Jul 29. Nat Rev Cancer. 2024. PMID: 39075222 Review.
References
-
- Andrews S. A quality control tool for high throughput sequence data. GPL v3Fastqc. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

