USP11 controls R-loops by regulating senataxin proteostasis
- PMID: 34526504
- PMCID: PMC8443744
- DOI: 10.1038/s41467-021-25459-w
USP11 controls R-loops by regulating senataxin proteostasis
Abstract
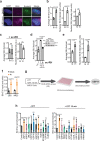
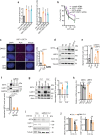
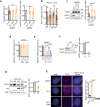
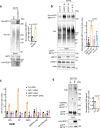
R-loops are by-products of transcription that must be tightly regulated to maintain genomic stability and gene expression. Here, we describe a mechanism for the regulation of the R-loop-specific helicase, senataxin (SETX), and identify the ubiquitin specific peptidase 11 (USP11) as an R-loop regulator. USP11 de-ubiquitinates SETX and its depletion increases SETX K48-ubiquitination and protein turnover. Loss of USP11 decreases SETX steady-state levels and reduces R-loop dissolution. Ageing of USP11 knockout cells restores SETX levels via compensatory transcriptional downregulation of the E3 ubiquitin ligase, KEAP1. Loss of USP11 reduces SETX enrichment at KEAP1 promoter, leading to R-loop accumulation, enrichment of the endonuclease XPF and formation of double-strand breaks. Overexpression of KEAP1 increases SETX K48-ubiquitination, promotes its degradation and R-loop accumulation. These data define a ubiquitination-dependent mechanism for SETX regulation, which is controlled by the opposing activities of USP11 and KEAP1 with broad applications for cancer and neurological disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Mutation in senataxin alters the mechanism of R-loop resolution in amyotrophic lateral sclerosis 4.Brain. 2022 Sep 14;145(9):3072-3094. doi: 10.1093/brain/awab464. Brain. 2022. PMID: 35045161 Free PMC article.
-
Senataxin RNA/DNA helicase promotes replication restart at co-transcriptional R-loops to prevent MUS81-dependent fork degradation.Nucleic Acids Res. 2024 Sep 23;52(17):10355-10369. doi: 10.1093/nar/gkae673. Nucleic Acids Res. 2024. PMID: 39119900 Free PMC article.
-
SETX (senataxin), the helicase mutated in AOA2 and ALS4, functions in autophagy regulation.Autophagy. 2021 Aug;17(8):1889-1906. doi: 10.1080/15548627.2020.1796292. Epub 2020 Aug 7. Autophagy. 2021. PMID: 32686621 Free PMC article.
-
Senataxin: A key actor in RNA metabolism, genome integrity and neurodegeneration.Biochimie. 2024 Feb;217:10-19. doi: 10.1016/j.biochi.2023.08.001. Epub 2023 Aug 7. Biochimie. 2024. PMID: 37558082 Review.
-
SUMOylated Senataxin functions in genome stability, RNA degradation, and stress granule disassembly, and is linked with inherited ataxia and motor neuron disease.Mol Genet Genomic Med. 2021 Dec;9(12):e1745. doi: 10.1002/mgg3.1745. Epub 2021 Jul 14. Mol Genet Genomic Med. 2021. PMID: 34263556 Free PMC article. Review.
Cited by
-
Beyond rRNA: nucleolar transcription generates a complex network of RNAs with multiple roles in maintaining cellular homeostasis.Genes Dev. 2022 Aug 1;36(15-16):876-886. doi: 10.1101/gad.349969.122. Genes Dev. 2022. PMID: 36207140 Free PMC article. Review.
-
C16orf72/HAPSTR1/TAPR1 functions with BRCA1/Senataxin to modulate replication-associated R-loops and confer resistance to PARP disruption.Nat Commun. 2023 Aug 17;14(1):5003. doi: 10.1038/s41467-023-40779-9. Nat Commun. 2023. PMID: 37591890 Free PMC article.
-
p53-dependent R-loop formation and HPV pathogenesis.Proc Natl Acad Sci U S A. 2023 Aug 29;120(35):e2305907120. doi: 10.1073/pnas.2305907120. Epub 2023 Aug 23. Proc Natl Acad Sci U S A. 2023. PMID: 37611058 Free PMC article.
-
Dysregulation of R-loop homeostasis shapes the immunosuppressive microenvironment and induces malignant progression in melanoma.Apoptosis. 2025 Feb;30(1-2):131-148. doi: 10.1007/s10495-024-02039-z. Epub 2024 Nov 2. Apoptosis. 2025. PMID: 39487313
-
Ubiquitin-specific protease 11 Aggravates Ischemia-reperfusion-induced Cardiomyocyte Pyroptosis and Injury by Promoting TRAF3 Deubiquitination.Balkan Med J. 2023 May 8;40(3):205-214. doi: 10.4274/balkanmedj.galenos.2023.2022-12-15. Epub 2023 Mar 31. Balkan Med J. 2023. PMID: 37000116 Free PMC article.
References
-
- Marinello J, Chillemi G, Bueno S, Manzo SG, Capranico G. Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res. 2013;41:10110–10123. doi: 10.1093/nar/gkt778. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

