SRGN-Triggered Aggressive and Immunosuppressive Phenotype in a Subset of TTF-1-Negative Lung Adenocarcinomas
- PMID: 34524427
- PMCID: PMC8826620
- DOI: 10.1093/jnci/djab183
SRGN-Triggered Aggressive and Immunosuppressive Phenotype in a Subset of TTF-1-Negative Lung Adenocarcinomas
Abstract
Background: Approximately 20% of lung adenocarcinoma (LUAD) is negative for the lineage-specific oncogene Thyroid transcription factor 1 (TTF-1) and exhibits worse clinical outcome with a low frequency of actionable genomic alterations. To identify molecular features associated with TTF-1-negative LUAD, we compared the transcriptomic and proteomic profiles of LUAD cell lines. SRGN , a chondroitin sulfate proteoglycan Serglycin, was identified as a markedly overexpressed gene in TTF-1-negative LUAD. We therefore investigated the roles and regulation of SRGN in TTF-1-negative LUAD.
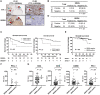
Methods: Proteomic and metabolomic analyses of 41 LUAD cell lines were done using mass spectrometry. The function of SRGN was investigated in 3 TTF-1-negative and 4 TTF-1-positive LUAD cell lines and in a syngeneic mouse model (n = 5 to 8 mice per group). Expression of SRGN was evaluated in 94 and 105 surgically resected LUAD tumor specimens using immunohistochemistry. All statistical tests were 2-sided.
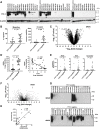
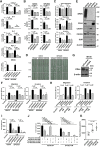
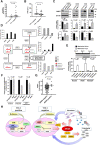
Results: SRGN was markedly overexpressed at mRNA and protein levels in TTF-1-negative LUAD cell lines (P < .001 for both mRNA and protein levels). Expression of SRGN in LUAD tumor tissue was associated with poor outcome (hazard ratio = 4.22, 95% confidence interval = 1.12 to 15.86, likelihood ratio test, P = .03), and with higher expression of Programmed cell death 1 ligand 1 (PD-L1) in tumor cells and higher infiltration of Programmed cell death protein 1-positive lymphocytes. SRGN regulated expression of PD-L1 as well as proinflammatory cytokines, including Interleukin-6, Interleukin-8, and C-X-C motif chemokine 1 in LUAD cell lines; increased migratory and invasive properties of LUAD cells and fibroblasts; and enhanced angiogenesis. SRGN was induced by DNA demethylation resulting from Nicotinamide N-methyltransferase-mediated impairment of methionine metabolism.
Conclusions: Our findings suggest that SRGN plays a pivotal role in tumor-stromal interaction and reprogramming into an aggressive and immunosuppressive tumor microenvironment in TTF-1-negative LUAD.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner.Oncogene. 2017 Apr 27;36(17):2457-2471. doi: 10.1038/onc.2016.404. Epub 2016 Nov 7. Oncogene. 2017. PMID: 27819672 Free PMC article.
-
Proteoglycan serglycin promotes non-small cell lung cancer cell migration through the interaction of its glycosaminoglycans with CD44.J Biomed Sci. 2020 Jan 2;27(1):2. doi: 10.1186/s12929-019-0600-3. J Biomed Sci. 2020. PMID: 31898491 Free PMC article.
-
Cancer-associated fibroblasts secrete hypoxia-induced serglycin to promote head and neck squamous cell carcinoma tumor cell growth in vitro and in vivo by activating the Wnt/β-catenin pathway.Cell Oncol (Dordr). 2021 Jun;44(3):661-671. doi: 10.1007/s13402-021-00592-2. Epub 2021 Mar 2. Cell Oncol (Dordr). 2021. PMID: 33651283
-
NKX2-1/TTF-1: an enigmatic oncogene that functions as a double-edged sword for cancer cell survival and progression.Cancer Cell. 2013 Jun 10;23(6):718-23. doi: 10.1016/j.ccr.2013.04.002. Cancer Cell. 2013. PMID: 23763999 Review.
-
Clinicopathological analysis of bronchiolar adenoma combined with lung adenocarcinoma: Report of eight cases and literature review.Histol Histopathol. 2024 Jun;39(6):783-794. doi: 10.14670/HH-18-682. Epub 2023 Nov 29. Histol Histopathol. 2024. PMID: 38059279 Review.
Cited by
-
Association of thyroid transcription factor-1 with the efficacy of immune-checkpoint inhibitors in patients with advanced lung adenocarcinoma.Thorac Cancer. 2022 Aug;13(16):2309-2317. doi: 10.1111/1759-7714.14560. Epub 2022 Jul 8. Thorac Cancer. 2022. PMID: 35808895 Free PMC article.
-
Thyroid transcription factor-1 (TTF-1) expression and the efficacy of combination therapy with immune checkpoint inhibitors and cytotoxic chemotherapy in non-squamous non-small cell lung cancer.Transl Lung Cancer Res. 2023 Sep 28;12(9):1850-1861. doi: 10.21037/tlcr-23-331. Epub 2023 Sep 13. Transl Lung Cancer Res. 2023. PMID: 37854151 Free PMC article.
-
Kynureninase Upregulation Is a Prominent Feature of NFR2-Activated Cancers and Is Associated with Tumor Immunosuppression and Poor Prognosis.Cancers (Basel). 2023 Jan 29;15(3):834. doi: 10.3390/cancers15030834. Cancers (Basel). 2023. PMID: 36765792 Free PMC article.
-
Metabolic barriers in non-small cell lung cancer with LKB1 and/or KEAP1 mutations for immunotherapeutic strategies.Front Oncol. 2023 Aug 22;13:1249237. doi: 10.3389/fonc.2023.1249237. eCollection 2023. Front Oncol. 2023. PMID: 37675220 Free PMC article. Review.
-
Relationship Between TTF-1 Expression and PFS of Pemetrexed-containing Chemotherapy in Non-squamous-NSCLC Patients With and Without Driver Genes.Cancer Diagn Progn. 2023 Jan 3;3(1):53-60. doi: 10.21873/cdp.10179. eCollection 2023 Jan-Feb. Cancer Diagn Progn. 2023. PMID: 36632586 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

