Neutralizing Monoclonal Antibodies That Target the Spike Receptor Binding Domain Confer Fc Receptor-Independent Protection against SARS-CoV-2 Infection in Syrian Hamsters
- PMID: 34517754
- PMCID: PMC8546861
- DOI: 10.1128/mBio.02395-21
Neutralizing Monoclonal Antibodies That Target the Spike Receptor Binding Domain Confer Fc Receptor-Independent Protection against SARS-CoV-2 Infection in Syrian Hamsters
Abstract
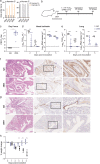
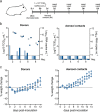
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein is the main target for neutralizing antibodies. These antibodies can be elicited through immunization or passively transferred as therapeutics in the form of convalescent-phase sera or monoclonal antibodies (MAbs). Potently neutralizing antibodies are expected to confer protection; however, it is unclear whether weakly neutralizing antibodies contribute to protection. Also, their mechanism of action in vivo is incompletely understood. Here, we demonstrate that 2B04, an antibody with an ultrapotent neutralizing activity (50% inhibitory concentration [IC50] of 0.04 μg/ml), protects hamsters against SARS-CoV-2 in a prophylactic and therapeutic infection model. Protection is associated with reduced weight loss and viral loads in nasal turbinates and lungs after challenge. MAb 2B04 also blocked aerosol transmission of the virus to naive contacts. We next examined three additional MAbs (2C02, 2C03, and 2E06), recognizing distinct epitopes within the receptor binding domain of spike protein that possess either minimal (2C02 and 2E06, IC50 > 20 μg/ml) or weak (2C03, IC50 of 5 μg/ml) virus neutralization capacity in vitro. Only 2C03 protected Syrian hamsters from weight loss and reduced lung viral load after SARS-CoV-2 infection. Finally, we demonstrated that Fc-Fc receptor interactions were not required for protection when 2B04 and 2C03 were administered prophylactically. These findings inform the mechanism of protection and support the rational development of antibody-mediated protection against SARS-CoV-2 infections. IMPORTANCE The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by SARS-CoV-2, has resulted in the loss of millions of lives. Safe and effective vaccines are considered the ultimate remedy for the global social and economic disruption caused by the pandemic. However, a thorough understanding of the immune correlates of protection against this virus is lacking. Here, we characterized four different monoclonal antibodies and evaluated their ability to prevent or treat SARS-CoV-2 infection in Syrian hamsters. These antibodies varied in their ability to neutralize the virus in vitro. Prophylactic administration of potent and weakly neutralizing antibodies protected against SARS-CoV-2 infection, and this effect was Fc receptor independent. The potent neutralizing antibody also had therapeutic efficacy and eliminated onward aerosol transmission. In contrast, minimally neutralizing antibodies provided no protection against infection with SARS-CoV-2 in Syrian hamsters. Combined, these studies highlight the significance of weakly neutralizing antibodies in the protection against SARS-CoV-2 infection and associated disease.
Keywords: COVID-19; SARS-CoV-2; Syrian hamster; monoclonal antibodies; transmission.
Figures



Similar articles
-
Immunogenicity and efficacy of XBB.1.5 rS vaccine against the EG.5.1 variant of SARS-CoV-2 in Syrian hamsters.J Virol. 2024 Oct 22;98(10):e0052824. doi: 10.1128/jvi.00528-24. Epub 2024 Sep 4. J Virol. 2024. PMID: 39230305
-
Highly Neutralizing COVID-19 Convalescent Plasmas Potently Block SARS-CoV-2 Replication and Pneumonia in Syrian Hamsters.J Virol. 2022 Feb 23;96(4):e0155121. doi: 10.1128/JVI.01551-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34818068 Free PMC article.
-
Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.J Virol. 2018 Apr 27;92(10):e02002-17. doi: 10.1128/JVI.02002-17. Print 2018 May 15. J Virol. 2018. PMID: 29514901 Free PMC article.
-
Protective non-neutralizing SARS-CoV-2 monoclonal antibodies.Trends Immunol. 2024 Aug;45(8):609-624. doi: 10.1016/j.it.2024.06.003. Epub 2024 Jul 20. Trends Immunol. 2024. PMID: 39034185 Review.
-
SARS-CoV-2 Neutralizing Antibodies for COVID-19 Prevention and Treatment.Annu Rev Med. 2022 Jan 27;73:1-16. doi: 10.1146/annurev-med-042420-113838. Epub 2021 Aug 24. Annu Rev Med. 2022. PMID: 34428080 Review.
Cited by
-
Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice.Sci Transl Med. 2022 May 25;14(646):eabn1252. doi: 10.1126/scitranslmed.abn1252. Epub 2022 May 25. Sci Transl Med. 2022. PMID: 35412328 Free PMC article.
-
Imprinting of serum neutralizing antibodies by Wuhan-1 mRNA vaccines.Nature. 2024 Jun;630(8018):950-960. doi: 10.1038/s41586-024-07539-1. Epub 2024 May 15. Nature. 2024. PMID: 38749479 Free PMC article.
-
Enhanced RBD-Specific Antibody Responses and SARS-CoV-2-Relevant T-Cell Activity in Healthcare Workers Following Booster Vaccination.Curr Issues Mol Biol. 2024 Oct 2;46(10):11124-11135. doi: 10.3390/cimb46100660. Curr Issues Mol Biol. 2024. PMID: 39451540 Free PMC article.
-
Antibody-mediated neutralization of SARS-CoV-2.Immunity. 2022 Jun 14;55(6):925-944. doi: 10.1016/j.immuni.2022.05.005. Epub 2022 May 13. Immunity. 2022. PMID: 35623355 Free PMC article. Review.
-
Safety and Immunogenicity of a Booster Vaccination by CoronaVac or BNT162b2 in Previously Two-Dose Inactivated Virus Vaccinated Individuals with Negative Neutralizing Antibody.Vaccines (Basel). 2022 Apr 3;10(4):556. doi: 10.3390/vaccines10040556. Vaccines (Basel). 2022. PMID: 35455305 Free PMC article.
References
-
- Barnes CO, West AP, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB, Gaebler C, Muecksch F, Lorenzi JCC, Finkin S, Hägglöf T, Hurley A, Millard KG, Weisblum Y, Schmidt F, Hatziioannou T, Bieniasz PD, Caskey M, Robbiani DF, Nussenzweig MC, Bjorkman PJ. 2020. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182:828–842.e16. doi:10.1016/j.cell.2020.06.025. - DOI - PMC - PubMed
-
- Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, Chai X, He R, Li X, Lv Q, Zhu H, Deng W, Xu Y, Wang Y, Qiao L, Tan Y, Song L, Wang G, Du X, Gao N, Liu J, Xiao J, Su X-D, Du Z, Feng Y, Qin C, Qin C, Jin R, Xie XS. 2020. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell 182:73–84 e16. doi:10.1016/j.cell.2020.05.025. - DOI - PMC - PubMed
-
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W. 2020. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science 369:650–655. doi:10.1126/science.abc6952. - DOI - PMC - PubMed
-
- Du S, Cao Y, Zhu Q, Yu P, Qi F, Wang G, Du X, Bao L, Deng W, Zhu H, Liu J, Nie J, Zheng Y, Liang H, Liu R, Gong S, Xu H, Yisimayi A, Lv Q, Wang B, He R, Han Y, Zhao W, Bai Y, Qu Y, Gao X, Ji C, Wang Q, Gao N, Huang W, Wang Y, Xie XS, Su X-D, Xiao J, Qin C. 2020. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell 183:1013–1023.e13. doi:10.1016/j.cell.2020.09.035. - DOI - PMC - PubMed
-
- Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. doi:10.1038/s41586-020-2380-z. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI151810/AI/NIAID NIH HHS/United States
- HHSN272201400006C/AI/NIAID NIH HHS/United States
- T11-712/19-N/Research Grants Council, University Grants Committee ()
- AI139251/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- COVID190126/FHB | Health and Medical Research Fund (HMRF)
- 75N93019C00051/AI/NIAID NIH HHS/United States
- AI151810-01/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI118938/AI/NIAID NIH HHS/United States
- U01 AI150747/AI/NIAID NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- U01 AI141990/AI/NIAID NIH HHS/United States
- R01 AI139251/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
