Single-Molecule Insights Into the Dynamics of Replicative Helicases
- PMID: 34513934
- PMCID: PMC8426354
- DOI: 10.3389/fmolb.2021.741718
Single-Molecule Insights Into the Dynamics of Replicative Helicases
Abstract
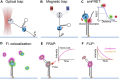
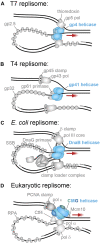
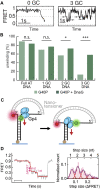
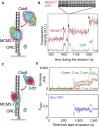
Helicases are molecular motors that translocate along single-stranded DNA and unwind duplex DNA. They rely on the consumption of chemical energy from nucleotide hydrolysis to drive their translocation. Specialized helicases play a critically important role in DNA replication by unwinding DNA at the front of the replication fork. The replicative helicases of the model systems bacteriophages T4 and T7, Escherichia coli and Saccharomyces cerevisiae have been extensively studied and characterized using biochemical methods. While powerful, their averaging over ensembles of molecules and reactions makes it challenging to uncover information related to intermediate states in the unwinding process and the dynamic helicase interactions within the replisome. Here, we describe single-molecule methods that have been developed in the last few decades and discuss the new details that these methods have revealed about replicative helicases. Applying methods such as FRET and optical and magnetic tweezers to individual helicases have made it possible to access the mechanistic aspects of unwinding. It is from these methods that we understand that the replicative helicases studied so far actively translocate and then passively unwind DNA, and that these hexameric enzymes must efficiently coordinate the stepping action of their subunits to achieve unwinding, where the size of each step is prone to variation. Single-molecule fluorescence microscopy methods have made it possible to visualize replicative helicases acting at replication forks and quantify their dynamics using multi-color colocalization, FRAP and FLIP. These fluorescence methods have made it possible to visualize helicases in replication initiation and dissect this intricate protein-assembly process. In a similar manner, single-molecule visualization of fluorescent replicative helicases acting in replication identified that, in contrast to the replicative polymerases, the helicase does not exchange. Instead, the replicative helicase acts as the stable component that serves to anchor the other replication factors to the replisome.
Keywords: DNA replication; dynamics; fluorescence; helicases; multi-protein complexes; replisome; single-molecule.
Copyright © 2021 Spinks, Spenkelink, Dixon and van Oijen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Single-Molecule FRET Analysis of Replicative Helicases.Methods Mol Biol. 2018;1805:233-250. doi: 10.1007/978-1-4939-8556-2_12. Methods Mol Biol. 2018. PMID: 29971721
-
Single-molecule perspectives on helicase mechanisms and functions.Crit Rev Biochem Mol Biol. 2016;51(1):15-25. doi: 10.3109/10409238.2015.1102195. Epub 2015 Nov 5. Crit Rev Biochem Mol Biol. 2016. PMID: 26540349 Review.
-
Methods to study the coupling between replicative helicase and leading-strand DNA polymerase at the replication fork.Methods. 2016 Oct 1;108:65-78. doi: 10.1016/j.ymeth.2016.05.003. Epub 2016 May 9. Methods. 2016. PMID: 27173619 Free PMC article. Review.
-
The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates.J Biol Chem. 2006 Sep 15;281(37):26914-21. doi: 10.1074/jbc.M605518200. Epub 2006 Jul 7. J Biol Chem. 2006. PMID: 16829518
-
Escherichia coli DNA helicases: mechanisms of DNA unwinding.Mol Microbiol. 1992 Jan;6(1):5-14. doi: 10.1111/j.1365-2958.1992.tb00831.x. Mol Microbiol. 1992. PMID: 1310794 Review.
Cited by
-
Assembly, Activation, and Helicase Actions of MCM2-7: Transition from Inactive MCM2-7 Double Hexamers to Active Replication Forks.Biology (Basel). 2024 Aug 17;13(8):629. doi: 10.3390/biology13080629. Biology (Basel). 2024. PMID: 39194567 Free PMC article. Review.
-
Observing protein dynamics during DNA-lesion bypass by the replisome.Front Mol Biosci. 2022 Sep 21;9:968424. doi: 10.3389/fmolb.2022.968424. eCollection 2022. Front Mol Biosci. 2022. PMID: 36213113 Free PMC article. Review.
-
Synergism between CMG helicase and leading strand DNA polymerase at replication fork.Nat Commun. 2023 Sep 20;14(1):5849. doi: 10.1038/s41467-023-41506-0. Nat Commun. 2023. PMID: 37730685 Free PMC article.
-
Episomal and chromosomal DNA replication and recombination in Entamoeba histolytica.Front Mol Biosci. 2023 Jun 8;10:1212082. doi: 10.3389/fmolb.2023.1212082. eCollection 2023. Front Mol Biosci. 2023. PMID: 37363402 Free PMC article. Review.
-
Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches.Int J Mol Sci. 2023 Feb 1;24(3):2806. doi: 10.3390/ijms24032806. Int J Mol Sci. 2023. PMID: 36769124 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

