Therapeutic Potential of Targeting the SUMO Pathway in Cancer
- PMID: 34503213
- PMCID: PMC8431684
- DOI: 10.3390/cancers13174402
Therapeutic Potential of Targeting the SUMO Pathway in Cancer
Abstract
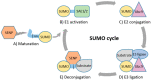
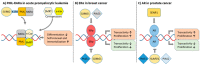
SUMOylation is a dynamic and reversible post-translational modification, characterized more than 20 years ago, that regulates protein function at multiple levels. Key oncoproteins and tumor suppressors are SUMO substrates. In addition to alterations in SUMO pathway activity due to conditions typically present in cancer, such as hypoxia, the SUMO machinery components are deregulated at the genomic level in cancer. The delicate balance between SUMOylation and deSUMOylation is regulated by SENP enzymes possessing SUMO-deconjugation activity. Dysregulation of SUMO machinery components can disrupt the balance of SUMOylation, contributing to the tumorigenesis and drug resistance of various cancers in a context-dependent manner. Many molecular mechanisms relevant to the pathogenesis of specific cancers involve SUMO, highlighting the potential relevance of SUMO machinery components as therapeutic targets. Recent advances in the development of inhibitors targeting SUMOylation and deSUMOylation permit evaluation of the therapeutic potential of targeting the SUMO pathway in cancer. Finally, the first drug inhibiting SUMO pathway, TAK-981, is currently also being evaluated in clinical trials in cancer patients. Intriguingly, the inhibition of SUMOylation may also have the potential to activate the anti-tumor immune response. Here, we comprehensively and systematically review the recent developments in understanding the role of SUMOylation in cancer and specifically focus on elaborating the scientific rationale of targeting the SUMO pathway in different cancers.
Keywords: cancer; post-translational modification (PTM); protein inhibitor of activated STAT (PIAS); sentrin-specific protease (SENP); small ubiquitin-like modifier (SUMO).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Regulation of ALS-Associated SOD1 Mutant SUMOylation and Aggregation by SENP and PIAS Family Proteins.J Mol Neurosci. 2020 Dec;70(12):2007-2014. doi: 10.1007/s12031-020-01604-w. Epub 2020 May 27. J Mol Neurosci. 2020. PMID: 32462635
-
Progress in the Discovery of Small Molecule Modulators of DeSUMOylation.Curr Issues Mol Biol. 2020;35:17-34. doi: 10.21775/cimb.035.017. Epub 2019 Aug 18. Curr Issues Mol Biol. 2020. PMID: 31422931 Review.
-
SUMOylation and DeSUMOylation: Prospective therapeutic targets in cancer.Life Sci. 2023 Nov 1;332:122085. doi: 10.1016/j.lfs.2023.122085. Epub 2023 Sep 16. Life Sci. 2023. PMID: 37722589 Review.
-
Discovery of a Dual SENP1 and SENP2 Inhibitor.Int J Mol Sci. 2022 Oct 11;23(20):12085. doi: 10.3390/ijms232012085. Int J Mol Sci. 2022. PMID: 36292935 Free PMC article.
-
Cancer-Associated Dysregulation of Sumo Regulators: Proteases and Ligases.Int J Mol Sci. 2022 Jul 20;23(14):8012. doi: 10.3390/ijms23148012. Int J Mol Sci. 2022. PMID: 35887358 Free PMC article. Review.
Cited by
-
dsRNAi-mediated silencing of PIAS2beta specifically kills anaplastic carcinomas by mitotic catastrophe.Nat Commun. 2024 May 14;15(1):3736. doi: 10.1038/s41467-024-47751-1. Nat Commun. 2024. PMID: 38744818 Free PMC article.
-
Protein Stability Regulation in Osteosarcoma: The Ubiquitin-like Modifications and Glycosylation as Mediators of Tumor Growth and as Targets for Therapy.Cells. 2024 Mar 18;13(6):537. doi: 10.3390/cells13060537. Cells. 2024. PMID: 38534381 Free PMC article. Review.
-
The Ubiquitin-Proteasome System in Tumor Metabolism.Cancers (Basel). 2023 Apr 20;15(8):2385. doi: 10.3390/cancers15082385. Cancers (Basel). 2023. PMID: 37190313 Free PMC article. Review.
-
Mechanistic update of Trisenox in blood cancer.Curr Res Pharmacol Drug Discov. 2023 Nov 20;5:100166. doi: 10.1016/j.crphar.2023.100166. eCollection 2023. Curr Res Pharmacol Drug Discov. 2023. PMID: 38074774 Free PMC article. Review.
-
Cardiac-targeted PIASy gene silencing mediates deSUMOylation of caveolin-3 and prevents ischemia/reperfusion-induced Nav1.5 downregulation and ventricular arrhythmias.Mil Med Res. 2022 Oct 14;9(1):58. doi: 10.1186/s40779-022-00415-x. Mil Med Res. 2022. PMID: 36229865 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

