Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs
- PMID: 34502324
- PMCID: PMC8431113
- DOI: 10.3390/ijms22179416
Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs
Abstract
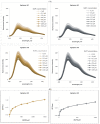
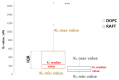
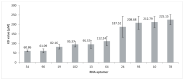
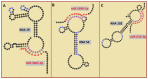
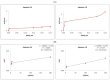
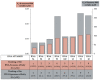
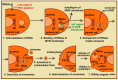
RNA motifs may promote interactions with exosomes (EXO-motifs) and lipid rafts (RAFT-motifs) that are enriched in exosomal membranes. These interactions can promote selective RNA loading into exosomes. We quantified the affinity between RNA aptamers containing various EXO- and RAFT-motifs and membrane lipid rafts in a liposome model of exosomes by determining the dissociation constants. Analysis of the secondary structure of RNA molecules provided data about the possible location of EXO- and RAFT-motifs within the RNA structure. The affinity of RNAs containing RAFT-motifs (UUGU, UCCC, CUCC, CCCU) and some EXO-motifs (CCCU, UCCU) to rafted liposomes is higher in comparison to aptamers without these motifs, suggesting direct RNA-exosome interaction. We have confirmed these results through the determination of the dissociation constant values of exosome-RNA aptamer complexes. RNAs containing EXO-motifs GGAG or UGAG have substantially lower affinity to lipid rafts, suggesting indirect RNA-exosome interaction via RNA binding proteins. Bioinformatics analysis revealed RNA aptamers containing both raft- and miRNA-binding motifs and involvement of raft-binding motifs UCCCU and CUCCC. A strategy is proposed for using functional RNA aptamers (fRNAa) containing both RAFT-motif and a therapeutic motif (e.g., miRNA inhibitor) to selectively introduce RNAs into exosomes for fRNAa delivery to target cells for personalized therapy.
Keywords: FRET spectroscopy; RNA aptamers; RNA motifs; exosomes; liposomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Binding of RNA Aptamers to Membrane Lipid Rafts: Implications for Exosomal miRNAs Transfer from Cancer to Immune Cells.Int J Mol Sci. 2020 Nov 12;21(22):8503. doi: 10.3390/ijms21228503. Int J Mol Sci. 2020. PMID: 33198080 Free PMC article.
-
Selection of bifunctional RNAs with specificity for arginine and lipid membranes.FEBS Lett. 2024 May;598(9):1061-1079. doi: 10.1002/1873-3468.14880. Epub 2024 Apr 22. FEBS Lett. 2024. PMID: 38649155
-
Mechanisms of RNA loading into exosomes.FEBS Lett. 2015 Jun 4;589(13):1391-8. doi: 10.1016/j.febslet.2015.04.036. Epub 2015 Apr 30. FEBS Lett. 2015. PMID: 25937124 Review.
-
Lipid Rafts in Exosome Biogenesis.Biochemistry (Mosc). 2020 Feb;85(2):177-191. doi: 10.1134/S0006297920020054. Biochemistry (Mosc). 2020. PMID: 32093594 Review.
-
Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles.BMC Genomics. 2011 Nov 30;12 Suppl 3(Suppl 3):S18. doi: 10.1186/1471-2164-12-S3-S18. Epub 2011 Nov 30. BMC Genomics. 2011. PMID: 22369587 Free PMC article.
Cited by
-
Exosomal cargos-mediated metabolic reprogramming in tumor microenvironment.J Exp Clin Cancer Res. 2023 Mar 10;42(1):59. doi: 10.1186/s13046-023-02634-z. J Exp Clin Cancer Res. 2023. PMID: 36899389 Free PMC article. Review.
-
Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges.Int J Mol Sci. 2023 Apr 14;24(8):7287. doi: 10.3390/ijms24087287. Int J Mol Sci. 2023. PMID: 37108446 Free PMC article. Review.
-
Exploring the roles and molecular mechanisms of RNA binding proteins in the sorting of noncoding RNAs into exosomes during tumor progression.J Adv Res. 2024 Nov;65:105-123. doi: 10.1016/j.jare.2023.11.029. Epub 2023 Nov 27. J Adv Res. 2024. PMID: 38030125 Free PMC article. Review.
-
Extracellular Vesicle-Based Method for Detecting MYCN Amplification Status of Pediatric Neuroblastoma.Cancers (Basel). 2022 May 26;14(11):2627. doi: 10.3390/cancers14112627. Cancers (Basel). 2022. PMID: 35681607 Free PMC article.
-
Extracellular vesicles could be a putative posttranscriptional regulatory mechanism that shapes intracellular RNA levels in Plasmodium falciparum.Nat Commun. 2023 Oct 13;14(1):6447. doi: 10.1038/s41467-023-42103-x. Nat Commun. 2023. PMID: 37833314 Free PMC article.
References
-
- Théry C., Regnault A., Garin J., Wolfers J., Zitvogel L., Ricciardi-Castagnoli P., Raposo G., Amigorena S. Molecular characterization of dendritic cell-derived exosomes: Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

