A New Multiplex Genetic Detection Assay Method for the Rapid Semi-Quantitative Detection of Six Common Curable Sexually Transmitted Pathogens From the Genital Tract
- PMID: 34497776
- PMCID: PMC8420868
- DOI: 10.3389/fcimb.2021.704037
A New Multiplex Genetic Detection Assay Method for the Rapid Semi-Quantitative Detection of Six Common Curable Sexually Transmitted Pathogens From the Genital Tract
Abstract
Background: Sexually transmitted infections (STIs) are some of the most common communicable conditions and exert impact on the health and lives of many hundreds of millions of people across the world every year. Screening high-risk populations and conducting comprehensive detection tests would lead to a significant improvement in preventing the transmission of STIs and help us to provide rapid treatment to those affected. Here, we successfully established and validated a novel high-throughput multiplex gene detection system (HMGS) for the simultaneous and semiquantitative detection of six important curable sexually transmitted pathogens in a single reaction from secretions samples.
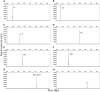
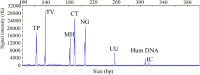
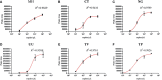
Method: Fluorescently labeled primers were designed to target specific conserved and single-copy gene fragments of Ureaplasma urealyticum (U. urealyticum), Mycoplasma hominis (M. hominis), Chlamydia trachomatis (C. trachomatis), Neisseria gonorrhoeae (N. gonorrhoeae), Trichomonas vaginalis (T. vaginalis), and Treponema pallidum (T. pallidum). The specificity and sensitivity of the STI-HMGS was validated and optimized using plasmids and quantitative genomic DNA. Next, we validated the performances of the STI-HMGS for clinical application by testing samples of clinical secretions collected from patients who visited the gynecology and urology outpatient clinics of our reproductive medicine center. Results derived from the STI-HMGS were then compared with three approved commercialized kits that used to detect U. urealyticum, C. trachomatis and N. gonorrhoeae, respectively, followed by further validation with Sanger sequencing for all pathogens. Finally, a comprehensive analysis of epidemiology was performed among different subgroups to investigate the association between infection rates and clinically-relevant information.
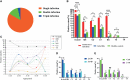
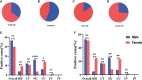
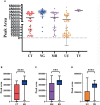
Results: The sensitivity of STI-HMGS for six target genes was 10 copies/µL. Data derived from the detection of 381 clinical secretions demonstrated that the STI-HMGS exhibited high concordance rate compared with approved commercialized kits and almost 100% sensitivity and specificity for the detection of six sexually transmitted pathogens when validated by Sanger sequencing. Semi-quantitative analysis found that STIs caused by N. gonorrhoeae had a significantly higher (P<0.05) pathogen load than the other pathogens. Infections caused by C. trachomatis were significantly more common in younger individuals (P<0.05). We also found that U. urealyticum infections were more likely to happen in females; while the males were more affected by N. gonorrhoeae (P<0.05).
Conclusions: STI-HMGS proved to be an efficient method for the semi-quantitative detection of six important curable sexually transmitted pathogens and therefore represents an alternative method for the clinical detection and monitoring of STIs.
Keywords: High-throughput multiplex gene detection system (HMGS); Sexually transmitted infections (STIs); pathogens; positive rates; rapid; semi-quantitative detection.
Copyright © 2021 Sun, Meng, Wang, Yang, liu, Zeng, Zhang, Zhu, Chi, Liu, Jiang, Ding, Miao, Wu, Zhao and Zhang.
Conflict of interest statement
Authors XZ, DZ, HWZ, and YW were employed by company Ningbo HEALTH Gene Technologies Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Clinical performance of four multiplex real-time PCR kits detecting urogenital and sexually transmitted pathogens.Clin Microbiol Infect. 2022 May;28(5):733.e7-733.e13. doi: 10.1016/j.cmi.2021.09.028. Epub 2021 Oct 2. Clin Microbiol Infect. 2022. PMID: 34610459
-
Cervical Cytology of Samples with Ureaplasma urealyticum, Ureaplasma parvum, Chlamydia trachomatis, Trichomonas vaginalis, Mycoplasma hominis, and Neisseria gonorrhoeae Detected by Multiplex PCR.Biomed Res Int. 2020 Jul 7;2020:7045217. doi: 10.1155/2020/7045217. eCollection 2020. Biomed Res Int. 2020. PMID: 32724807 Free PMC article.
-
Highly specific and efficient primers for in-house multiplex PCR detection of Chlamydia trachomatis, Neisseria gonorrhoeae, Mycoplasma hominis and Ureaplasma urealyticum.BMC Res Notes. 2014 Jul 6;7:433. doi: 10.1186/1756-0500-7-433. BMC Res Notes. 2014. PMID: 24997675 Free PMC article.
-
Should we be testing for urogenital Mycoplasma hominis, Ureaplasma parvum and Ureaplasma urealyticum in men and women? - a position statement from the European STI Guidelines Editorial Board.J Eur Acad Dermatol Venereol. 2018 Nov;32(11):1845-1851. doi: 10.1111/jdv.15146. Epub 2018 Jul 6. J Eur Acad Dermatol Venereol. 2018. PMID: 29924422 Review.
-
Advances in the Understanding and Treatment of Male Urethritis.Clin Infect Dis. 2015 Dec 15;61 Suppl 8:S763-9. doi: 10.1093/cid/civ755. Clin Infect Dis. 2015. PMID: 26602615 Review.
Cited by
-
Clinical characteristics of the host DNA-removed metagenomic next-generation sequencing technology for detecting SARS-CoV-2, revealing host local immune signaling and assisting genomic epidemiology.Front Immunol. 2022 Nov 15;13:1016440. doi: 10.3389/fimmu.2022.1016440. eCollection 2022. Front Immunol. 2022. PMID: 36458015 Free PMC article.
-
Detection of Ureaplasma urealyticum by Catalytic Hairpin Assembly Combined with a Lateral Flow Immunoassay Strip.ACS Omega. 2022 Sep 15;7(38):33830-33836. doi: 10.1021/acsomega.2c02457. eCollection 2022 Sep 27. ACS Omega. 2022. PMID: 36188314 Free PMC article.
-
Rapid and simultaneous detection of multiple pathogens in the lower reproductive tract during pregnancy based on loop-mediated isothermal amplification-microfluidic chip.BMC Microbiol. 2022 Oct 29;22(1):260. doi: 10.1186/s12866-022-02657-0. BMC Microbiol. 2022. PMID: 36309654 Free PMC article.
-
Copan Walk Away Specimen Processor (WASP) Automated System for Pathogen Detection in Female Reproductive Tract Specimens.Front Cell Infect Microbiol. 2021 Nov 17;11:770367. doi: 10.3389/fcimb.2021.770367. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34869072 Free PMC article.
References
-
- Bittencourt M. D. J. S., Brito A. C. D., Nascimento B. A. M. D., Carvalho A. H., Nascimento M. D. D. (2015). A Case of Secondary Syphilis Mimicking Palmoplantar Psoriasis in HIV Infected Patient. Anais brasileiros dermatología. 90 (3 Suppl 1), S216–S219. 10.1590/abd1806-4841.20153399 - DOI - PMC - PubMed
-
- Carnicer-Pont D., Loureiro-Varela E., Manresa J. M., Martinez M., Avecilla À., Montero-Pons L., et al. . (2019). The Notijoves Project: Protocol for a Randomized Controlled Trial About New Communication Technologies and Gamification to Promote Partner Notification of Sexually Transmitted Infections Among Young People. JMIR Res. Protoc. 8 (6), e12896. 10.2196/12896 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

