SLX4IP N-terminus dictates telomeric localization in ALT-like castration-resistant prostate cancer cell lines
- PMID: 34492133
- PMCID: PMC8460604
- DOI: 10.1002/pros.24225
SLX4IP N-terminus dictates telomeric localization in ALT-like castration-resistant prostate cancer cell lines
Abstract
Background: To ensure replicative immortality in cancer, telomeres must be maintained through activation of telomere maintenance mechanisms (TMMs) that are dependent on telomerase or the alternative lengthening of telomeres (ALT) pathway. Although TMM pathways have traditionally been considered to be mutually exclusive, ALT hallmarks have been identified in cancers defined as being telomerase-positive, supporting TMM coexistence. In castration-resistant prostate cancer (CRPC), in vitro models were thought to be universally dependent on telomerase as the primary TMM; however, CRPC models with androgen receptor (AR) loss demonstrate ALT hallmarks with limited telomerase activity and require ALT-associated PML bodies (APBs) for sustained telomere maintenance. The TMM coexistence in AR-negative CRPC is reliant on the ALT regulator protein, SLX4IP.
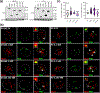
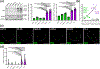
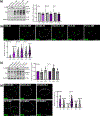
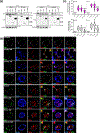
Methods: To identify the regions of SLX4IP responsible for the induction of APBs and telomere preservation in CRPC models, five 3xFLAG-tagged SLX4IP constructs were designed and stably introduced into parental C4-2B, DU145, and PC-3 cells. Once generated, these cell lines were interrogated for APB abundance and SLX4IP construct localization via immunofluorescence-fluorescence in situ hybridization (IF-FISH) and coimmunoprecipitation experiments for telomeric localization. Similarly, PC-3 cells with endogenous SLX4IP knockdown and SLX4IP construct introduction were interrogated for APB abundance, telomere length preservation, and senescent rescue.
Results: Here, we define the N-terminus of SLX4IP as being responsible for the promotion of the ALT-like phenotype of AR-negative CRPC models. Specifically, the N-terminus of SLX4IP was sufficient for promoting APB formation to a similar degree as full-length SLX4IP across CRPC cell lines. Additionally, APB promotion by the N-terminus of SLX4IP rescued telomere shortening and senescent induction triggered by SLX4IP knockdown in AR-negative CRPC cells. Moreover, APB formation and telomere maintenance were dependent on the ability of the N-terminus to direct SLX4IP localization at telomeres and APBs.
Conclusions: These findings identify the role of the uncharacterized ALT regulator SLX4IP in the promotion of TMM coexistence to perpetuate replicative immortality in CRPC in vitro.
Keywords: ALT-associated PML body; PML; androgen receptor-negative prostate cancer; androgen-independent prostate cancer; telomere maintenance mechanism.
© 2021 Wiley Periodicals LLC.
Figures



Similar articles
-
SLX4IP Promotes Telomere Maintenance in Androgen Receptor-Independent Castration-Resistant Prostate Cancer through ALT-like Telomeric PML Localization.Mol Cancer Res. 2021 Feb;19(2):301-316. doi: 10.1158/1541-7786.MCR-20-0314. Epub 2020 Nov 13. Mol Cancer Res. 2021. PMID: 33188147 Free PMC article.
-
Telomere length heterogeneity in ALT cells is maintained by PML-dependent localization of the BTR complex to telomeres.Genes Dev. 2020 May 1;34(9-10):650-662. doi: 10.1101/gad.333963.119. Epub 2020 Mar 26. Genes Dev. 2020. PMID: 32217664 Free PMC article.
-
Telomere maintenance in Wilms tumors: first evidence for the presence of alternative lengthening of telomeres mechanism.Genes Chromosomes Cancer. 2011 Oct;50(10):823-9. doi: 10.1002/gcc.20903. Epub 2011 Jul 18. Genes Chromosomes Cancer. 2011. PMID: 21769957
-
Telomere Length Maintenance in Cancer: At the Crossroad between Telomerase and Alternative Lengthening of Telomeres (ALT).Int J Mol Sci. 2018 Feb 18;19(2):606. doi: 10.3390/ijms19020606. Int J Mol Sci. 2018. PMID: 29463031 Free PMC article. Review.
-
ALT: A Multi-Faceted Phenomenon.Genes (Basel). 2020 Jan 27;11(2):133. doi: 10.3390/genes11020133. Genes (Basel). 2020. PMID: 32012790 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

