Elicitation of Broadly Neutralizing Antibodies against B.1.1.7, B.1.351, and B.1.617.1 SARS-CoV-2 Variants by Three Prototype Strain-Derived Recombinant Protein Vaccines
- PMID: 34452287
- PMCID: PMC8402859
- DOI: 10.3390/v13081421
Elicitation of Broadly Neutralizing Antibodies against B.1.1.7, B.1.351, and B.1.617.1 SARS-CoV-2 Variants by Three Prototype Strain-Derived Recombinant Protein Vaccines
Abstract
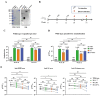
The ongoing coronavirus disease 2019 (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Most of the currently approved SARS-CoV-2 vaccines use the prototype strain-derived spike (S) protein or its receptor-binding domain (RBD) as the vaccine antigen. The emergence of several novel SARS-CoV-2 variants has raised concerns about potential immune escape. In this study, we performed an immunogenicity comparison of prototype strain-derived RBD, S1, and S ectodomain trimer (S-trimer) antigens and evaluated their induction of neutralizing antibodies against three circulating SARS-CoV-2 variants, including B.1.1.7, B.1.351, and B.1.617.1. We found that, at the same antigen dose, the RBD and S-trimer vaccines were more potent than the S1 vaccine in eliciting long-lasting, high-titer broadly neutralizing antibodies in mice. The RBD immune sera remained highly effective against the B.1.1.7, B.1.351, and B.1.617.1 variants despite the corresponding neutralizing titers decreasing by 1.2-, 2.8-, and 3.5-fold relative to that against the wild-type strain. Significantly, the S-trimer immune sera exhibited comparable neutralization potency (less than twofold variation in neutralizing GMTs) towards the prototype strain and all three variants tested. These findings provide valuable information for further development of recombinant protein-based SARS-CoV-2 vaccines and support the continued use of currently approved SARS-CoV-2 vaccines in the regions/countries where variant viruses circulate.
Keywords: COVID-19; S1; SARS-CoV-2 variants; neutralizing antibody; receptor binding domain; spike protein; vaccine.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Pan-beta-coronavirus subunit vaccine prevents SARS-CoV-2 Omicron, SARS-CoV, and MERS-CoV challenge.J Virol. 2024 Sep 17;98(9):e0037624. doi: 10.1128/jvi.00376-24. Epub 2024 Aug 27. J Virol. 2024. PMID: 39189731
-
Glycan Masking of Epitopes in the NTD and RBD of the Spike Protein Elicits Broadly Neutralizing Antibodies Against SARS-CoV-2 Variants.Front Immunol. 2021 Dec 2;12:795741. doi: 10.3389/fimmu.2021.795741. eCollection 2021. Front Immunol. 2021. PMID: 34925381 Free PMC article.
-
Immunogenicity and efficacy of XBB.1.5 rS vaccine against the EG.5.1 variant of SARS-CoV-2 in Syrian hamsters.J Virol. 2024 Oct 22;98(10):e0052824. doi: 10.1128/jvi.00528-24. Epub 2024 Sep 4. J Virol. 2024. PMID: 39230305
-
Comprehensive Overview of Broadly Neutralizing Antibodies against SARS-CoV-2 Variants.Viruses. 2024 Jun 1;16(6):900. doi: 10.3390/v16060900. Viruses. 2024. PMID: 38932192 Free PMC article. Review.
-
Variants of SARS-CoV-2, their effects on infection, transmission and neutralization by vaccine-induced antibodies.Eur Rev Med Pharmacol Sci. 2021 Sep;25(18):5857-5864. doi: 10.26355/eurrev_202109_26805. Eur Rev Med Pharmacol Sci. 2021. PMID: 34604978 Review.
Cited by
-
SARS-CoV-2 Variants, Current Vaccines and Therapeutic Implications for COVID-19.Vaccines (Basel). 2022 Sep 16;10(9):1538. doi: 10.3390/vaccines10091538. Vaccines (Basel). 2022. PMID: 36146616 Free PMC article. Review.
-
Human Fc-Conjugated Receptor Binding Domain-Based Recombinant Subunit Vaccines with Short Linker Induce Potent Neutralizing Antibodies against Multiple SARS-CoV-2 Variants.Vaccines (Basel). 2022 Sep 8;10(9):1502. doi: 10.3390/vaccines10091502. Vaccines (Basel). 2022. PMID: 36146579 Free PMC article.
-
The Variation of SARS-CoV-2 and Advanced Research on Current Vaccines.Front Med (Lausanne). 2022 Jan 18;8:806641. doi: 10.3389/fmed.2021.806641. eCollection 2021. Front Med (Lausanne). 2022. PMID: 35118097 Free PMC article. Review.
-
Human Organoids and Organs-on-Chips for Addressing COVID-19 Challenges.Adv Sci (Weinh). 2022 Apr;9(10):e2105187. doi: 10.1002/advs.202105187. Epub 2022 Feb 2. Adv Sci (Weinh). 2022. PMID: 35107217 Free PMC article. Review.
-
Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.Emerg Microbes Infect. 2022 Dec;11(1):351-367. doi: 10.1080/22221751.2021.2024455. Emerg Microbes Infect. 2022. PMID: 34964428 Free PMC article.
References
-
- Xu C., Wang Y.X., Liu C.X., Zhang C., Han W.Y., Hong X.Y., Wang Y.F., Hong Q., Wang S.T., Zhao Q.Y., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021;7:eabe5575. doi: 10.1126/sciadv.abe5575. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

