AKTIP interacts with ESCRT I and is needed for the recruitment of ESCRT III subunits to the midbody
- PMID: 34449766
- PMCID: PMC8428793
- DOI: 10.1371/journal.pgen.1009757
AKTIP interacts with ESCRT I and is needed for the recruitment of ESCRT III subunits to the midbody
Abstract
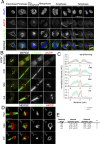
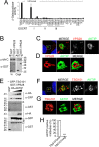
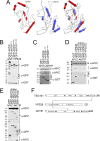
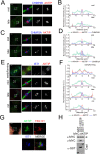
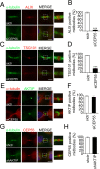
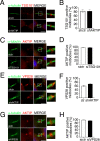
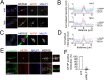
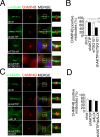
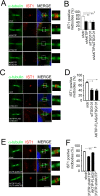
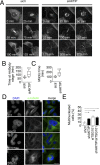
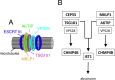
To complete mitosis, the bridge that links the two daughter cells needs to be cleaved. This step is carried out by the endosomal sorting complex required for transport (ESCRT) machinery. AKTIP, a protein discovered to be associated with telomeres and the nuclear membrane in interphase cells, shares sequence similarities with the ESCRT I component TSG101. Here we present evidence that during mitosis AKTIP is part of the ESCRT machinery at the midbody. AKTIP interacts with the ESCRT I subunit VPS28 and forms a circular supra-structure at the midbody, in close proximity with TSG101 and VPS28 and adjacent to the members of the ESCRT III module CHMP2A, CHMP4B and IST1. Mechanistically, the recruitment of AKTIP is dependent on MKLP1 and independent of CEP55. AKTIP and TSG101 are needed together for the recruitment of the ESCRT III subunit CHMP4B and in parallel for the recruitment of IST1. Alone, the reduction of AKTIP impinges on IST1 and causes multinucleation. Our data altogether reveal that AKTIP is a component of the ESCRT I module and functions in the recruitment of ESCRT III components required for abscission.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
A Septin Double Ring Controls the Spatiotemporal Organization of the ESCRT Machinery in Cytokinetic Abscission.Curr Biol. 2019 Jul 8;29(13):2174-2182.e7. doi: 10.1016/j.cub.2019.05.050. Epub 2019 Jun 13. Curr Biol. 2019. PMID: 31204162 Free PMC article.
-
Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4846-51. doi: 10.1073/pnas.1102714108. Epub 2011 Mar 7. Proc Natl Acad Sci U S A. 2011. PMID: 21383202 Free PMC article.
-
UMAD1 contributes to ESCRT-III dynamic subunit turnover during cytokinetic abscission.J Cell Sci. 2023 Aug 1;136(15):jcs261097. doi: 10.1242/jcs.261097. Epub 2023 Aug 10. J Cell Sci. 2023. PMID: 37439191 Free PMC article.
-
ESCRT machinery and cytokinesis: the road to daughter cell separation.Traffic. 2011 Oct;12(10):1318-26. doi: 10.1111/j.1600-0854.2011.01244.x. Epub 2011 Jul 27. Traffic. 2011. PMID: 21722282 Review.
-
ESCRT function in cytokinesis: location, dynamics and regulation by mitotic kinases.Int J Mol Sci. 2014 Nov 25;15(12):21723-39. doi: 10.3390/ijms151221723. Int J Mol Sci. 2014. PMID: 25429432 Free PMC article. Review.
Cited by
-
Phase Separation in the Nucleus and at the Nuclear Periphery during Post-Mitotic Nuclear Envelope Reformation.Cells. 2022 May 25;11(11):1749. doi: 10.3390/cells11111749. Cells. 2022. PMID: 35681444 Free PMC article. Review.
-
Mechanics and regulation of cytokinetic abscission.Front Cell Dev Biol. 2022 Nov 24;10:1046617. doi: 10.3389/fcell.2022.1046617. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506096 Free PMC article. Review.
-
Amorphous silica nanoparticles cause abnormal cytokinesis and multinucleation through dysfunction of the centralspindlin complex and microfilaments.Part Fibre Toxicol. 2023 Aug 22;20(1):34. doi: 10.1186/s12989-023-00544-8. Part Fibre Toxicol. 2023. PMID: 37608338 Free PMC article.
-
Impact of diffused versus vasculature targeted DNA damage on the heart of mice depleted of telomeric factor Ft1.Aging Cell. 2023 Dec;22(12):e14022. doi: 10.1111/acel.14022. Epub 2023 Nov 13. Aging Cell. 2023. PMID: 37960940 Free PMC article.
-
Close Ties between the Nuclear Envelope and Mammalian Telomeres: Give Me Shelter.Genes (Basel). 2023 Mar 23;14(4):775. doi: 10.3390/genes14040775. Genes (Basel). 2023. PMID: 37107534 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

