Emerging Single-Cell Technological Approaches to Investigate Chromatin Dynamics and Centromere Regulation in Human Health and Disease
- PMID: 34445507
- PMCID: PMC8395756
- DOI: 10.3390/ijms22168809
Emerging Single-Cell Technological Approaches to Investigate Chromatin Dynamics and Centromere Regulation in Human Health and Disease
Abstract
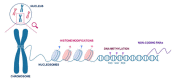
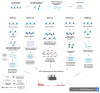
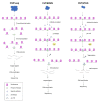
Epigenetic regulators play a crucial role in establishing and maintaining gene expression states. To date, the main efforts to study cellular heterogeneity have focused on elucidating the variable nature of the chromatin landscape. Specific chromatin organisation is fundamental for normal organogenesis and developmental homeostasis and can be affected by different environmental factors. The latter can lead to detrimental alterations in gene transcription, as well as pathological conditions such as cancer. Epigenetic marks regulate the transcriptional output of cells. Centromeres are chromosome structures that are epigenetically regulated and are crucial for accurate segregation. The advent of single-cell epigenetic profiling has provided finer analytical resolution, exposing the intrinsic peculiarities of different cells within an apparently homogenous population. In this review, we discuss recent advances in methodologies applied to epigenetics, such as CUT&RUN and CUT&TAG. Then, we compare standard and emerging single-cell techniques and their relevance for investigating human diseases. Finally, we describe emerging methodologies that investigate centromeric chromatin specification and neocentromere formation.
Keywords: centromere; chromatin regulation; emerging technologies; epigenetics; epigenetics in human health & disease; single-cell epigenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Centromeric chromatin and the pathway that drives its propagation.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):313-21. Biochim Biophys Acta. 2013. PMID: 24459733 Review.
-
Switching the centromeres on and off: epigenetic chromatin alterations provide plasticity in centromere activity stabilizing aberrant dicentric chromosomes.Biochem Soc Trans. 2013 Dec;41(6):1648-53. doi: 10.1042/BST20130136. Biochem Soc Trans. 2013. PMID: 24256269 Review.
-
Microtubule dynamics decoded by the epigenetic state of centromeric chromatin.Curr Genet. 2016 Nov;62(4):691-695. doi: 10.1007/s00294-016-0588-0. Epub 2016 Mar 14. Curr Genet. 2016. PMID: 26976145 Review.
-
Genetic and epigenetic effects on centromere establishment.Chromosoma. 2020 Mar;129(1):1-24. doi: 10.1007/s00412-019-00727-3. Epub 2019 Nov 28. Chromosoma. 2020. PMID: 31781852 Review.
-
A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance.J Cell Biol. 2015 Jun 22;209(6):789-801. doi: 10.1083/jcb.201503132. Epub 2015 Jun 15. J Cell Biol. 2015. PMID: 26076692 Free PMC article.
Cited by
-
The CUT&RUN greenlist: genomic regions of consistent noise are effective normalizing factors for quantitative epigenome mapping.Brief Bioinform. 2024 Jan 22;25(2):bbad538. doi: 10.1093/bib/bbad538. Brief Bioinform. 2024. PMID: 38279652 Free PMC article.
-
Deciphering the molecular basis of tissue-specific gene expression in plants: Can synthetic biology help?Curr Opin Plant Biol. 2022 Aug;68:102241. doi: 10.1016/j.pbi.2022.102241. Epub 2022 Jun 11. Curr Opin Plant Biol. 2022. PMID: 35700675 Free PMC article. Review.
-
The fly homolog of SUPT16H, a gene associated with neurodevelopmental disorders, is required in a cell-autonomous fashion for cell survival.Hum Mol Genet. 2023 Mar 6;32(6):984-997. doi: 10.1093/hmg/ddac259. Hum Mol Genet. 2023. PMID: 36255738 Free PMC article.
-
From DNA Copy Number Gains and Tumor Dependencies to Novel Therapeutic Targets for High-Risk Neuroblastoma.J Pers Med. 2021 Dec 3;11(12):1286. doi: 10.3390/jpm11121286. J Pers Med. 2021. PMID: 34945759 Free PMC article. Review.
-
Single-nucleotide resolution detection of Topo IV cleavage activity in the Escherichia coli genome with Topo-Seq.Front Microbiol. 2023 Apr 6;14:1160736. doi: 10.3389/fmicb.2023.1160736. eCollection 2023. Front Microbiol. 2023. PMID: 37089538 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

