The Changes in the p53 Protein across the Animal Kingdom Point to Its Involvement in Longevity
- PMID: 34445220
- PMCID: PMC8395165
- DOI: 10.3390/ijms22168512
The Changes in the p53 Protein across the Animal Kingdom Point to Its Involvement in Longevity
Abstract
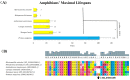
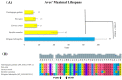
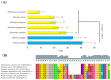
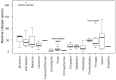
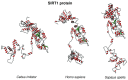
Recently, the quest for the mythical fountain of youth has produced extensive research programs that aim to extend the healthy lifespan of humans. Despite advances in our understanding of the aging process, the surprisingly extended lifespan and cancer resistance of some animal species remain unexplained. The p53 protein plays a crucial role in tumor suppression, tissue homeostasis, and aging. Long-lived, cancer-free African elephants have 20 copies of the TP53 gene, including 19 retrogenes (38 alleles), which are partially active, whereas humans possess only one copy of TP53 and have an estimated cancer mortality rate of 11-25%. The mechanism through which p53 contributes to the resolution of the Peto's paradox in Animalia remains vague. Thus, in this work, we took advantage of the available datasets and inspected the p53 amino acid sequence of phylogenetically related organisms that show variations in their lifespans. We discovered new correlations between specific amino acid deviations in p53 and the lifespans across different animal species. We found that species with extended lifespans have certain characteristic amino acid substitutions in the p53 DNA-binding domain that alter its function, as depicted from the Phenotypic Annotation of p53 Mutations, using the PROVEAN tool or SWISS-MODEL workflow. In addition, the loop 2 region of the human p53 DNA-binding domain was identified as the longest region that was associated with longevity. The 3D model revealed variations in the loop 2 structure in long-lived species when compared with human p53. Our findings show a direct association between specific amino acid residues in p53 protein, changes in p53 functionality, and the extended animal lifespan, and further highlight the importance of p53 protein in aging.
Keywords: aging; comparative analysis; longevity; p53; protein sequence.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






Similar articles
-
Solutions to Peto's paradox revealed by mathematical modelling and cross-species cancer gene analysis.Philos Trans R Soc Lond B Biol Sci. 2015 Jul 19;370(1673):20140222. doi: 10.1098/rstb.2014.0222. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26056366 Free PMC article.
-
A polymorphism in the tumor suppressor p53 affects aging and longevity in mouse models.Elife. 2018 Mar 20;7:e34701. doi: 10.7554/eLife.34701. Elife. 2018. PMID: 29557783 Free PMC article.
-
The Role of TP53 in Adaptation and Evolution.Cells. 2023 Feb 3;12(3):512. doi: 10.3390/cells12030512. Cells. 2023. PMID: 36766853 Free PMC article. Review.
-
Reactivation of mutant p53: Constraints on mechanism highlighted by principal component analysis of the DNA binding domain.Proteins. 2016 Oct;84(10):1443-61. doi: 10.1002/prot.25089. Epub 2016 Jul 26. Proteins. 2016. PMID: 27317883
-
Aging: What We Can Learn From Elephants.Front Aging. 2021 Aug 26;2:726714. doi: 10.3389/fragi.2021.726714. eCollection 2021. Front Aging. 2021. PMID: 35822016 Free PMC article. Review.
Cited by
-
Impacts of Molecular Structure on Nucleic Acid-Protein Interactions.Int J Mol Sci. 2022 Dec 26;24(1):407. doi: 10.3390/ijms24010407. Int J Mol Sci. 2022. PMID: 36613851 Free PMC article.
-
Senescence program and its reprogramming in pancreatic premalignancy.Cell Death Dis. 2023 Aug 17;14(8):528. doi: 10.1038/s41419-023-06040-3. Cell Death Dis. 2023. PMID: 37591827 Free PMC article. Review.
-
Onco-Breastomics: An Eco-Evo-Devo Holistic Approach.Int J Mol Sci. 2024 Jan 28;25(3):1628. doi: 10.3390/ijms25031628. Int J Mol Sci. 2024. PMID: 38338903 Free PMC article. Review.
-
Thermal stress, p53 structures and learning from elephants.Cell Death Discov. 2024 Aug 7;10(1):353. doi: 10.1038/s41420-024-02109-w. Cell Death Discov. 2024. PMID: 39107279 Free PMC article. Review.
-
Circular RNAs Involved in the Regulation of the Age-Related Pathways.Int J Mol Sci. 2022 Sep 9;23(18):10443. doi: 10.3390/ijms231810443. Int J Mol Sci. 2022. PMID: 36142352 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

