The Role of microRNAs in NK Cell Development and Function
- PMID: 34440789
- PMCID: PMC8391642
- DOI: 10.3390/cells10082020
The Role of microRNAs in NK Cell Development and Function
Abstract
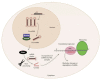
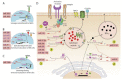
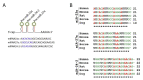
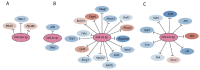
The clinical use of natural killer (NK) cells is at the forefront of cellular therapy. NK cells possess exceptional antitumor cytotoxic potentials and can generate significant levels of proinflammatory cytokines. Multiple genetic manipulations are being tested to augment the anti-tumor functions of NK cells. One such method involves identifying and altering microRNAs (miRNAs) that play essential roles in the development and effector functions of NK cells. Unique miRNAs can bind and inactivate mRNAs that code for cytotoxic proteins. MicroRNAs, such as the members of the Mirc11 cistron, downmodulate ubiquitin ligases that are central to the activation of the obligatory transcription factors responsible for the production of inflammatory cytokines. These studies reveal potential opportunities to post-translationally enhance the effector functions of human NK cells while reducing unwanted outcomes. Here, we summarize the recent advances made on miRNAs in murine and human NK cells and their relevance to NK cell development and functions.
Keywords: Mirc11; NK cells; inflammation; microRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Mirc11 Disrupts Inflammatory but Not Cytotoxic Responses of NK Cells.Cancer Immunol Res. 2019 Oct;7(10):1647-1662. doi: 10.1158/2326-6066.CIR-18-0934. Epub 2019 Sep 12. Cancer Immunol Res. 2019. PMID: 31515257 Free PMC article.
-
T-cells require post-transcriptional regulation for accurate immune responses.Biochem Soc Trans. 2015 Dec;43(6):1201-7. doi: 10.1042/BST20150154. Biochem Soc Trans. 2015. PMID: 26614661 Review.
-
Identification of microRNA transcriptome involved in human natural killer cell activation.Immunol Lett. 2012 Apr 30;143(2):208-17. doi: 10.1016/j.imlet.2012.02.014. Immunol Lett. 2012. PMID: 22701882
-
microRNA management of NK-cell developmental and functional programs.Eur J Immunol. 2014 Oct;44(10):2862-8. doi: 10.1002/eji.201444798. Epub 2014 Sep 16. Eur J Immunol. 2014. PMID: 25142111 Free PMC article. Review.
-
MicroRNA-15b Modulates Japanese Encephalitis Virus-Mediated Inflammation via Targeting RNF125.J Immunol. 2015 Sep 1;195(5):2251-62. doi: 10.4049/jimmunol.1500370. Epub 2015 Jul 22. J Immunol. 2015. PMID: 26202983
Cited by
-
The Regulatory Activity of Noncoding RNAs in ILCs.Cells. 2021 Oct 14;10(10):2742. doi: 10.3390/cells10102742. Cells. 2021. PMID: 34685721 Free PMC article. Review.
-
The New Kid on the Block: HLA-C, a Key Regulator of Natural Killer Cells in Viral Immunity.Cells. 2021 Nov 10;10(11):3108. doi: 10.3390/cells10113108. Cells. 2021. PMID: 34831331 Free PMC article. Review.
-
MicroRNAs: Small but Key Players in Viral Infections and Immune Responses to Viral Pathogens.Biology (Basel). 2023 Oct 14;12(10):1334. doi: 10.3390/biology12101334. Biology (Basel). 2023. PMID: 37887044 Free PMC article. Review.
-
The role of miRNAs in viral myocarditis, and its possible implication in induction of mRNA-based COVID-19 vaccines-induced myocarditis.Bull Natl Res Cent. 2022;46(1):267. doi: 10.1186/s42269-022-00955-1. Epub 2022 Nov 17. Bull Natl Res Cent. 2022. PMID: 36415483 Free PMC article. Review.
-
miR-142: A Master Regulator in Hematological Malignancies and Therapeutic Opportunities.Cells. 2023 Dec 30;13(1):84. doi: 10.3390/cells13010084. Cells. 2023. PMID: 38201290 Free PMC article. Review.
References
-
- Lanier L.L., Phillips J.H., Hackett J., Tutt M., Kumar V. Natural killer cells: Definition of a cell type rather than a function. J. Immunol. 1986;137:2735–2739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

