Circadian clock core component Bmal1 dictates cell cycle rhythm of proliferating hepatocytes during liver regeneration
- PMID: 34431407
- PMCID: PMC8560370
- DOI: 10.1152/ajpgi.00204.2021
Circadian clock core component Bmal1 dictates cell cycle rhythm of proliferating hepatocytes during liver regeneration
Abstract
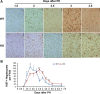
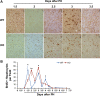
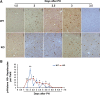
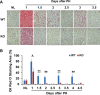
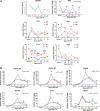
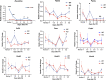
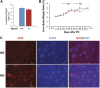
After partial hepatectomy (PH), the majority of remnant hepatocytes synchronously enter and rhythmically progress through the cell cycle for three major rounds to regain lost liver mass. Whether and how the circadian clock core component Bmal1 modulates this process remains elusive. We performed PH on Bmal1+/+ and hepatocyte-specific Bmal1 knockout (Bmal1hep-/-) mice and compared the initiation and progression of the hepatocyte cell cycle. After PH, Bmal1+/+ hepatocytes exhibited three major waves of nuclear DNA synthesis. In contrast, in Bmal1hep-/- hepatocytes, the first wave of nuclear DNA synthesis was delayed by 12 h, and the third such wave was lost. Following PH, Bmal1+/+ hepatocytes underwent three major waves of mitosis, whereas Bmal1hep-/- hepatocytes fully abolished mitotic oscillation. These Bmal1-dependent disruptions in the rhythmicity of hepatocyte cell cycle after PH were accompanied by suppressed expression peaks of a group of cell cycle components and regulators and dysregulated activation patterns of mitogenic signaling molecules c-Met and epidermal growth factor receptor. Moreover, Bmal1+/+ hepatocytes rhythmically accumulated fat as they expanded following PH, whereas this phenomenon was largely inhibited in Bmal1hep-/- hepatocytes. In addition, during late stages of liver regrowth, Bmal1 absence in hepatocytes caused the activation of redox sensor Nrf2, suggesting an oxidative stress state in regenerated liver tissue. Collectively, we demonstrated that during liver regeneration, Bmal1 partially modulates the oscillation of S-phase progression, fully controls the rhythmicity of M-phase advancement, and largely governs fluctuations in fat metabolism in replicating hepatocytes, as well as eventually determines the redox state of regenerated livers.NEW & NOTEWORTHY We demonstrated that Bmal1 centrally controls the synchronicity and rhythmicity of the cell cycle and lipid accumulation in replicating hepatocytes during liver regeneration. Bmal1 plays these roles, at least in part, by ensuring formation of the expression peaks of cell cycle components and regulators, as well as the timing and levels of activation of mitogenic signaling molecules.
Keywords: Bmal1; cell cycle; fat accumulation; hepatocyte proliferation; liver regeneration.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures

Similar articles
-
Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration.PLoS One. 2012;7(2):e30675. doi: 10.1371/journal.pone.0030675. Epub 2012 Feb 3. PLoS One. 2012. PMID: 22319576 Free PMC article.
-
Nrf2 is essential for timely M phase entry of replicating hepatocytes during liver regeneration.Am J Physiol Gastrointest Liver Physiol. 2015 Feb 15;308(4):G262-8. doi: 10.1152/ajpgi.00332.2014. Epub 2014 Dec 18. Am J Physiol Gastrointest Liver Physiol. 2015. PMID: 25524062 Free PMC article.
-
Keap1 modulates the redox cycle and hepatocyte cell cycle in regenerating liver.Cell Cycle. 2014;13(15):2349-58. doi: 10.4161/cc.29298. Cell Cycle. 2014. PMID: 25483186 Free PMC article.
-
Signal transduction during liver regeneration.J Gastroenterol Hepatol. 1998 Sep;13 Suppl:S93-5. J Gastroenterol Hepatol. 1998. PMID: 9792040 Review.
-
Neural function of Bmal1: an overview.Cell Biosci. 2023 Jan 2;13(1):1. doi: 10.1186/s13578-022-00947-8. Cell Biosci. 2023. PMID: 36593479 Free PMC article. Review.
Cited by
-
Chronobiology of Cancers in the Liver and Gut.Cancers (Basel). 2024 Aug 23;16(17):2925. doi: 10.3390/cancers16172925. Cancers (Basel). 2024. PMID: 39272783 Free PMC article. Review.
-
The circadian rhythm: an influential soundtrack in the diabetes story.Front Endocrinol (Lausanne). 2023 Jun 27;14:1156757. doi: 10.3389/fendo.2023.1156757. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37441501 Free PMC article. Review.
-
Endogenous Circadian Clock Machinery in Cortical NG2-Glia Regulates Cellular Proliferation.eNeuro. 2022 Oct 4;9(5):ENEURO.0110-22.2022. doi: 10.1523/ENEURO.0110-22.2022. Print 2022 Sep-Oct. eNeuro. 2022. PMID: 36123116 Free PMC article.
-
PPAR-γ Agonist Pioglitazone Restored Mouse Liver mRNA Expression of Clock Genes and Inflammation-Related Genes Disrupted by Reversed Feeding.PPAR Res. 2022 May 26;2022:7537210. doi: 10.1155/2022/7537210. eCollection 2022. PPAR Res. 2022. PMID: 35663475 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

