Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3)
- PMID: 34403472
- PMCID: PMC8464030
- DOI: 10.1093/nar/gkab712
Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3)
Abstract
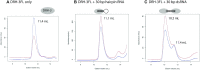
DRH-3 is critically involved in germline development and RNA interference (RNAi) facilitated chromosome segregation via the 22G-siRNA pathway in Caenorhabditis elegans. DRH-3 has similar domain architecture to RIG-I-like receptors (RLRs) and belongs to the RIG-I-like RNA helicase family. The molecular understanding of DRH-3 and its function in endogenous RNAi pathways remains elusive. In this study, we solved the crystal structures of the DRH-3 N-terminal domain (NTD) and the C-terminal domains (CTDs) in complex with 5'-triphosphorylated RNAs. The NTD of DRH-3 adopts a distinct fold of tandem caspase activation and recruitment domains (CARDs) structurally similar to the CARDs of RIG-I and MDA5, suggesting a signaling function in the endogenous RNAi biogenesis. The CTD preferentially recognizes 5'-triphosphorylated double-stranded RNAs bearing the typical features of secondary siRNA transcripts. The full-length DRH-3 displays unique structural dynamics upon binding to RNA duplexes that differ from RIG-I or MDA5. These features of DRH-3 showcase the evolutionary divergence of the Dicer and RLR family of helicases.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

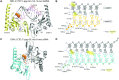


Similar articles
-
Caenorhabditis elegans RIG-I Homolog Mediates Antiviral RNA Interference Downstream of Dicer-Dependent Biogenesis of Viral Small Interfering RNAs.mBio. 2017 Mar 21;8(2):e00264-17. doi: 10.1128/mBio.00264-17. mBio. 2017. PMID: 28325765 Free PMC article.
-
Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA.Elife. 2024 May 15;13:RP93979. doi: 10.7554/eLife.93979. Elife. 2024. PMID: 38747717 Free PMC article.
-
Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms.Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):16085-90. doi: 10.1073/pnas.1307453110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043766 Free PMC article.
-
Structures of RIG-I-Like Receptors and Insights into Viral RNA Sensing.Adv Exp Med Biol. 2019;1172:157-188. doi: 10.1007/978-981-13-9367-9_8. Adv Exp Med Biol. 2019. PMID: 31628656 Review.
-
The molecular mechanism of RIG-I activation and signaling.Immunol Rev. 2021 Nov;304(1):154-168. doi: 10.1111/imr.13022. Epub 2021 Sep 12. Immunol Rev. 2021. PMID: 34514601 Free PMC article. Review.
Cited by
-
Creating de novo peptide-based bioactivities: from assembly to origami.RSC Adv. 2022 Sep 12;12(40):25955-25961. doi: 10.1039/d2ra03135c. eCollection 2022 Sep 12. RSC Adv. 2022. PMID: 36199601 Free PMC article. Review.
-
C. elegans RIG-I-like receptor DRH-1 signals via CARDs to activate anti-viral immunity in intestinal cells.bioRxiv [Preprint]. 2024 Feb 8:2024.02.05.578694. doi: 10.1101/2024.02.05.578694. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2402126121. doi: 10.1073/pnas.2402126121 PMID: 38370651 Free PMC article. Updated. Preprint.
-
A loosened gating mechanism of RIG-I leads to autoimmune disorders.Nucleic Acids Res. 2022 Jun 10;50(10):5850-5863. doi: 10.1093/nar/gkac361. Nucleic Acids Res. 2022. PMID: 35580046 Free PMC article.
-
Caenorhabditis elegans RIG-I-like receptor DRH-1 signals via CARDs to activate antiviral immunity in intestinal cells.Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2402126121. doi: 10.1073/pnas.2402126121. Epub 2024 Jul 9. Proc Natl Acad Sci U S A. 2024. PMID: 38980902 Free PMC article.
References
-
- Tabara H., Yigit E., Siomi H., Mello C.C.. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002; 109:861–871. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

