BiasNet: A Model to Predict Ligand Bias Toward GPCR Signaling
- PMID: 34397210
- PMCID: PMC8482801
- DOI: 10.1021/acs.jcim.1c00317
BiasNet: A Model to Predict Ligand Bias Toward GPCR Signaling
Abstract
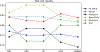
Signaling bias is a feature of many G protein-coupled receptor (GPCR) targeting drugs with potential clinical implications. Whether it is therapeutically advantageous for a drug to be G protein biased or β-arrestin biased depends on the context of the signaling pathway. Here, we explored GPCR ligands that exhibit biased signaling to gain insights into scaffolds and pharmacophores that lead to bias. More specifically, we considered BiasDB, a database containing information about GPCR biased ligands, and focused our analysis on ligands which show either a G protein or β-arrestin bias. Five different machine learning models were trained on these ligands using 15 different sets of features. Molecular fragments which were important for training the models were analyzed. Two of these fragments (number of secondary amines and number of aromatic amines) were more prevalent in β-arrestin biased ligands. After training a random forest model on HierS scaffolds, we found five scaffolds, which demonstrated G protein or β-arrestin bias. We also conducted t-SNE clustering, observing correspondence between unsupervised and supervised machine learning methods. To increase the applicability of our work, we developed a web implementation of our models, which can predict bias based on user-provided SMILES, drug names, or PubChem CID. Our web implementation is available at: drugdiscovery.utep.edu/biasnet.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
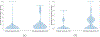


Similar articles
-
Screening β-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors.J Biomol Screen. 2013 Jun;18(5):599-609. doi: 10.1177/1087057113475480. Epub 2013 Feb 8. J Biomol Screen. 2013. PMID: 23396314
-
G-Protein/β-Arrestin-Linked Fluctuating Network of G-Protein-Coupled Receptors for Predicting Drug Efficacy and Bias Using Short-Term Molecular Dynamics Simulation.PLoS One. 2016 May 17;11(5):e0155816. doi: 10.1371/journal.pone.0155816. eCollection 2016. PLoS One. 2016. PMID: 27187591 Free PMC article.
-
Elucidating structural and molecular mechanisms of β-arrestin-biased agonism at GPCRs via MS-based proteomics.Cell Signal. 2018 Jan;41:56-64. doi: 10.1016/j.cellsig.2017.09.013. Epub 2017 Sep 20. Cell Signal. 2018. PMID: 28939107 Review.
-
A structural basis for how ligand binding site changes can allosterically regulate GPCR signaling and engender functional selectivity.Sci Signal. 2020 Feb 4;13(617):eaaw5885. doi: 10.1126/scisignal.aaw5885. Sci Signal. 2020. PMID: 32019899 Free PMC article.
-
Beta-arrestin-biased ligands at seven-transmembrane receptors.Trends Pharmacol Sci. 2007 Aug;28(8):416-22. doi: 10.1016/j.tips.2007.06.006. Epub 2007 Jul 20. Trends Pharmacol Sci. 2007. PMID: 17644195 Review.
Cited by
-
Epidermal growth factor receptor cascade prioritizes the maximization of signal transduction.Sci Rep. 2022 Oct 10;12(1):16950. doi: 10.1038/s41598-022-20663-0. Sci Rep. 2022. PMID: 36216834 Free PMC article.
-
New characterization of dihydroergotamine receptor pharmacology in the context of migraine: utilization of a β-arrestin recruitment assay.Front Neurol. 2023 Nov 21;14:1282846. doi: 10.3389/fneur.2023.1282846. eCollection 2023. Front Neurol. 2023. PMID: 38073648 Free PMC article.
-
Can machine learning 'transform' peptides/peptidomimetics into small molecules? A case study with ghrelin receptor ligands.Mol Divers. 2023 Oct;27(5):2239-2255. doi: 10.1007/s11030-022-10555-w. Epub 2022 Nov 4. Mol Divers. 2023. PMID: 36331785
-
GPCR-IPL score: multilevel featurization of GPCR-ligand interaction patterns and prediction of ligand functions from selectivity to biased activation.Brief Bioinform. 2024 Jan 22;25(2):bbae105. doi: 10.1093/bib/bbae105. Brief Bioinform. 2024. PMID: 38517694 Free PMC article.
-
Orphan G protein-coupled receptors: the ongoing search for a home.Front Pharmacol. 2024 Feb 29;15:1349097. doi: 10.3389/fphar.2024.1349097. eCollection 2024. Front Pharmacol. 2024. PMID: 38495099 Free PMC article. Review.
References
-
- Tan L; Yan W; McCorvy JD; Cheng J Biased Ligands of G Protein-Coupled Receptors (GPCRs): Structure-Functional Selectivity Relationships (SFSRs) and Therapeutic Potential. J. Med. Chem 2018, 61, 9841–9878. - PubMed
-
- Kenakin T Agonist-receptor efficacy II: agonist trafficking of receptor signals. Trends Pharmacol. Sci 1995, 16, 232–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

