Identification of candidate biomarkers correlated with poor prognosis of breast cancer based on bioinformatics analysis
- PMID: 34384030
- PMCID: PMC8806858
- DOI: 10.1080/21655979.2021.1960775
Identification of candidate biomarkers correlated with poor prognosis of breast cancer based on bioinformatics analysis
Abstract
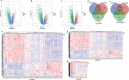
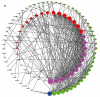
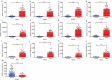
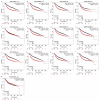
Breast cancer (BC) is a malignancy with high incidence among women in the world. This study aims to screen key genes and potential prognostic biomarkers for BC using bioinformatics analysis. Total 58 normal tissues and 203 cancer tissues were collected from three Gene Expression Omnibus (GEO) gene expression profiles, and then the differential expressed genes (DEGs) were identified. Subsequently, the Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway were analyzed to investigate the biological function of DEGs. Additionally, hub genes were screened by constructing a protein-protein interaction (PPI) network. Then, we explored the prognostic value and molecular mechanism of these hub genes using Kaplan-Meier (KM) curve and Gene Set Enrichment Analysis (GSEA). As a result, 42 up-regulated and 82 down-regulated DEGs were screened out from GEO datasets. The DEGs were mainly related to cell cycles and cell proliferation by GO and KEGG pathway analysis. Furthermore, 12 hub genes (FN1, AURKA, CCNB1, BUB1B, PRC1, TPX2, NUSAP1, TOP2A, KIF20A, KIF2C, RRM2, ASPM) with a high degree were identified initially, among which, 11 hub genes were significantly correlated with the prognosis of BC patients based on the Kaplan-Meier-plotter. GSEA reviewed that these hub genes correlated with KEGG_CELL_CYCLE and HALLMARK_P53_PATHWAY. In conclusion, this study identified 11 key genes as BC potential prognosis biomarkers on the basis of integrated bioinformatics analysis. This finding will improve our knowledge of the BC progress and mechanisms.
Keywords: Bioinformatics; breast cancer; gene expression omnibus; hub genes; prognosis biomarker.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis.Cell Transplant. 2019 Dec;28(1_suppl):76S-86S. doi: 10.1177/0963689719893950. Epub 2019 Dec 11. Cell Transplant. 2019. PMID: 31822116 Free PMC article.
-
Identification of potential key genes for HER-2 positive breast cancer based on bioinformatics analysis.Medicine (Baltimore). 2020 Jan;99(1):e18445. doi: 10.1097/MD.0000000000018445. Medicine (Baltimore). 2020. PMID: 31895772 Free PMC article.
-
Identification of crucial hub genes and potential molecular mechanisms in breast cancer by integrated bioinformatics analysis and experimental validation.Comput Biol Med. 2022 Oct;149:106036. doi: 10.1016/j.compbiomed.2022.106036. Epub 2022 Aug 25. Comput Biol Med. 2022. PMID: 36096037
-
Transcriptomic Signatures in Colorectal Cancer Progression.Curr Mol Med. 2023;23(3):239-249. doi: 10.2174/1566524022666220427102048. Curr Mol Med. 2023. PMID: 35490318 Review.
-
Exploring the role of estrogen and progestins in breast cancer: A genomic approach to diagnosis.Adv Protein Chem Struct Biol. 2024;142:25-43. doi: 10.1016/bs.apcsb.2023.12.023. Epub 2024 Jun 11. Adv Protein Chem Struct Biol. 2024. PMID: 39059987 Review.
Cited by
-
Doxorubicin downregulates cell cycle regulatory hub genes in breast cancer cells.Med Oncol. 2024 Aug 8;41(9):220. doi: 10.1007/s12032-024-02468-5. Med Oncol. 2024. PMID: 39115587
-
Differential gene expression analysis pipelines and bioinformatic tools for the identification of specific biomarkers: A review.Comput Struct Biotechnol J. 2024 Mar 1;23:1154-1168. doi: 10.1016/j.csbj.2024.02.018. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38510977 Free PMC article. Review.
-
A Case of Multi-Organ Tuberculosis Misdiagnosed as Lung Cancer and a Literature Review.Cancer Manag Res. 2023 Dec 27;15:1395-1400. doi: 10.2147/CMAR.S433226. eCollection 2023. Cancer Manag Res. 2023. PMID: 38161787 Free PMC article.
-
DLGAP5 Regulates the Proliferation, Migration, Invasion, and Cell Cycle of Breast Cancer Cells via the JAK2/STAT3 Signaling Axis.Int J Mol Sci. 2023 Oct 31;24(21):15819. doi: 10.3390/ijms242115819. Int J Mol Sci. 2023. PMID: 37958803 Free PMC article.
-
A Network of 17 Microtubule-Related Genes Highlights Functional Deregulations in Breast Cancer.Cancers (Basel). 2023 Oct 6;15(19):4870. doi: 10.3390/cancers15194870. Cancers (Basel). 2023. PMID: 37835564 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
