Single-molecule measurements reveal that PARP1 condenses DNA by loop stabilization
- PMID: 34380612
- PMCID: PMC8357241
- DOI: 10.1126/sciadv.abf3641
Single-molecule measurements reveal that PARP1 condenses DNA by loop stabilization
Abstract
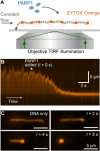
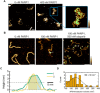
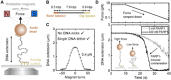
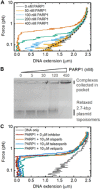
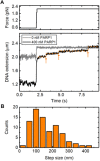
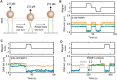
Poly(ADP-ribose) polymerase 1 (PARP1) is an abundant nuclear enzyme that plays important roles in DNA repair, chromatin organization and transcription regulation. Although binding and activation of PARP1 by DNA damage sites has been extensively studied, little is known about how PARP1 binds to long stretches of undamaged DNA and how it could shape chromatin architecture. Here, using single-molecule techniques, we show that PARP1 binds and condenses undamaged, kilobase-length DNA subject to sub-piconewton mechanical forces. Stepwise decondensation at high force and DNA braiding experiments show that the condensation activity is due to the stabilization of DNA loops by PARP1. PARP inhibitors do not affect the level of condensation of undamaged DNA but act to block condensation reversal for damaged DNA in the presence of NAD+ Our findings suggest a mechanism for PARP1 in the organization of chromatin structure.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures


Similar articles
-
Mechanistic insight into the role of Poly(ADP-ribosyl)ation in DNA topology modulation and response to DNA damage.Mutagenesis. 2020 Feb 13;35(1):107-118. doi: 10.1093/mutage/gez045. Mutagenesis. 2020. PMID: 31782485 Review.
-
Poly(ADP-ribose) Polymerase (PARP) and PARP Inhibitors: Mechanisms of Action and Role in Cardiovascular Disorders.Cardiovasc Toxicol. 2018 Dec;18(6):493-506. doi: 10.1007/s12012-018-9462-2. Cardiovasc Toxicol. 2018. PMID: 29968072 Review.
-
A Single-Molecule Atomic Force Microscopy Study of PARP1 and PARP2 Recognition of Base Excision Repair DNA Intermediates.J Mol Biol. 2019 Jul 12;431(15):2655-2673. doi: 10.1016/j.jmb.2019.05.028. Epub 2019 May 23. J Mol Biol. 2019. PMID: 31129062
-
Phosphatase 1 Nuclear Targeting Subunit Mediates Recruitment and Function of Poly (ADP-Ribose) Polymerase 1 in DNA Repair.Cancer Res. 2019 May 15;79(10):2526-2535. doi: 10.1158/0008-5472.CAN-18-1673. Epub 2019 Feb 7. Cancer Res. 2019. PMID: 30733193 Free PMC article.
-
MORC2 regulates DNA damage response through a PARP1-dependent pathway.Nucleic Acids Res. 2019 Sep 19;47(16):8502-8520. doi: 10.1093/nar/gkz545. Nucleic Acids Res. 2019. PMID: 31616951 Free PMC article.
Cited by
-
METTL3 promotes oxaliplatin resistance of gastric cancer CD133+ stem cells by promoting PARP1 mRNA stability.Cell Mol Life Sci. 2022 Feb 18;79(3):135. doi: 10.1007/s00018-022-04129-0. Cell Mol Life Sci. 2022. PMID: 35179655 Free PMC article.
-
ADP-ribosylation from molecular mechanisms to therapeutic implications.Cell. 2023 Oct 12;186(21):4475-4495. doi: 10.1016/j.cell.2023.08.030. Cell. 2023. PMID: 37832523 Free PMC article. Review.
-
PARP1 modulates METTL3 promoter chromatin accessibility and associated LPAR5 RNA m6A methylation to control cancer cell radiosensitivity.Mol Ther. 2023 Sep 6;31(9):2633-2650. doi: 10.1016/j.ymthe.2023.07.018. Epub 2023 Jul 23. Mol Ther. 2023. PMID: 37482682 Free PMC article.
-
Single-molecule force spectroscopy reveals binding and bridging dynamics of PARP1 and PARP2 at DNA double-strand breaks.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2214209120. doi: 10.1073/pnas.2214209120. Epub 2023 May 22. Proc Natl Acad Sci U S A. 2023. PMID: 37216533 Free PMC article.
-
Transcriptional regulation mechanism of PARP1 and its application in disease treatment.Epigenetics Chromatin. 2024 Aug 8;17(1):26. doi: 10.1186/s13072-024-00550-w. Epigenetics Chromatin. 2024. PMID: 39118189 Free PMC article. Review.
References
-
- Gibson B. A., Kraus W. L., New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012). - PubMed
-
- Benjamin R. C., Gill D. M., Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 255, 10502–10508 (1980). - PubMed
-
- Strickfaden H., McDonald D., Kruhlak M. J., Haince J.-F., Th’ng J. P. H., Rouleau M., Ishibashi T., Corry G. N., Ausio J., Underhill D. A., Poirier G. G., Hendzel M. J., Poly (ADP-ribosyl)ation-dependent transient chromatin decondensation and histone displacement following laser microirradiation. J. Biol. Chem. 291, 1789–1802 (2016). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

