Bacteriophage λ RexA and RexB functions assist the transition from lysogeny to lytic growth
- PMID: 34379857
- PMCID: PMC8541928
- DOI: 10.1111/mmi.14792
Bacteriophage λ RexA and RexB functions assist the transition from lysogeny to lytic growth
Abstract
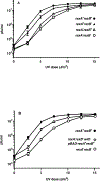
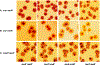
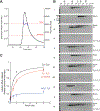
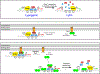
The CI and Cro repressors of bacteriophage λ create a bistable switch between lysogenic and lytic growth. In λ lysogens, CI repressor expressed from the PRM promoter blocks expression of the lytic promoters PL and PR to allow stable maintenance of the lysogenic state. When lysogens are induced, CI repressor is inactivated and Cro repressor is expressed from the lytic PR promoter. Cro repressor blocks PRM transcription and CI repressor synthesis to ensure that the lytic state proceeds. RexA and RexB proteins, like CI, are expressed from the PRM promoter in λ lysogens; RexB is also expressed from a second promoter, PLIT , embedded in rexA. Here we show that RexA binds CI repressor and assists the transition from lysogenic to lytic growth, using both intact lysogens and defective prophages with reporter genes under the control of the lytic PL and PR promoters. Once lytic growth begins, if the bistable switch does return to the immune state, RexA expression lessens the probability that it will remain there, thus stabilizing the lytic state and activation of the lytic PL and PR promoters. RexB modulates the effect of RexA and may also help establish phage DNA replication as lytic growth ensues.
Keywords: bacteriophage λ; genetic switch; lysogeny; lytic growth; phage development; prophage.
© 2021 John Wiley & Sons Ltd. This article has been contributed to by US Government employees and their work is in the public domain in the USA.
Figures
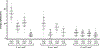
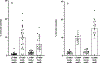
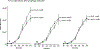



Similar articles
-
Elements in the λ immunity region regulate phage development: beyond the 'Genetic Switch'.Mol Microbiol. 2019 Dec;112(6):1798-1813. doi: 10.1111/mmi.14394. Epub 2019 Oct 8. Mol Microbiol. 2019. PMID: 31545538 Free PMC article.
-
Polarity within pM and pE promoted phage lambda cI-rexA-rexB transcription and its suppression.Can J Microbiol. 2005 Jan;51(1):37-49. doi: 10.1139/w04-115. Can J Microbiol. 2005. PMID: 15782233
-
Over-expression of rexA nullifies T4rII exclusion in Escherichia coli K(lambda) lysogens.Can J Microbiol. 2004 Feb;50(2):133-6. doi: 10.1139/w03-115. Can J Microbiol. 2004. PMID: 15052316
-
Yet another way that phage λ manipulates its Escherichia coli host: λrexB is involved in the lysogenic-lytic switch.Mol Microbiol. 2015 May;96(4):689-93. doi: 10.1111/mmi.12969. Epub 2015 Mar 16. Mol Microbiol. 2015. PMID: 25684601 Review.
-
Switches in bacteriophage lambda development.Annu Rev Genet. 2005;39:409-29. doi: 10.1146/annurev.genet.39.073003.113656. Annu Rev Genet. 2005. PMID: 16285866 Review.
Cited by
-
Recombineering: Genetic Engineering in Escherichia coli Using Homologous Recombination.Curr Protoc. 2023 Feb;3(2):e656. doi: 10.1002/cpz1.656. Curr Protoc. 2023. PMID: 36779782 Free PMC article.
-
The microbiome-product colibactin hits unique cellular targets mediating host-microbe interaction.Front Pharmacol. 2022 Sep 12;13:958012. doi: 10.3389/fphar.2022.958012. eCollection 2022. Front Pharmacol. 2022. PMID: 36172175 Free PMC article. Review.
-
Study of Ren, RexA, and RexB Functions Provides Insight Into the Complex Interaction Between Bacteriophage λ and Its Host, Escherichia coli.Phage (New Rochelle). 2022 Sep 1;3(3):153-164. doi: 10.1089/phage.2022.0020. Epub 2022 Sep 19. Phage (New Rochelle). 2022. PMID: 36204488 Free PMC article.
-
Hybrid Vigor: Importance of Hybrid λ Phages in Early Insights in Molecular Biology.Microbiol Mol Biol Rev. 2022 Dec 21;86(4):e0012421. doi: 10.1128/mmbr.00124-21. Epub 2022 Oct 19. Microbiol Mol Biol Rev. 2022. PMID: 36165780 Free PMC article. Review.
-
The crystal structure of bacteriophage λ RexA provides novel insights into the DNA binding properties of Rex-like phage exclusion proteins.Nucleic Acids Res. 2024 May 8;52(8):4659-4675. doi: 10.1093/nar/gkae212. Nucleic Acids Res. 2024. PMID: 38554102 Free PMC article.
References
-
- Baek K, Svenningsen S, Eisen H, Sneppen K, & Brown S (2003). Single-cell analysis of λ immunity regulation. J Mol Biol, 334(3), 363–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

