Conversion of dietary inositol into propionate and acetate by commensal Anaerostipes associates with host health
- PMID: 34376656
- PMCID: PMC8355322
- DOI: 10.1038/s41467-021-25081-w
Conversion of dietary inositol into propionate and acetate by commensal Anaerostipes associates with host health
Abstract
We describe the anaerobic conversion of inositol stereoisomers to propionate and acetate by the abundant intestinal genus Anaerostipes. A inositol pathway was elucidated by nuclear magnetic resonance using [13C]-inositols, mass spectrometry and proteogenomic analyses in A. rhamnosivorans, identifying 3-oxoacid CoA transferase as a key enzyme involved in both 3-oxopropionyl-CoA and propionate formation. This pathway also allowed conversion of phytate-derived inositol into propionate as shown with [13C]-phytate in fecal samples amended with A. rhamnosivorans. Metabolic and (meta)genomic analyses explained the adaptation of Anaerostipes spp. to inositol-containing substrates and identified a propionate-production gene cluster to be inversely associated with metabolic biomarkers in (pre)diabetes cohorts. Co-administration of myo-inositol with live A. rhamnosivorans in western-diet fed mice reduced fasting-glucose levels comparing to heat-killed A. rhamnosivorans after 6-weeks treatment. Altogether, these data suggest a potential beneficial role for intestinal Anaerostipes spp. in promoting host health.
© 2021. The Author(s).
Conflict of interest statement
The Wageningen University has applied for a patent relating to the use of bacteria capable of converting inositol into propionate on which T.P.N.B. and W.M.dV. are inventors (Wageningen; WO2021028585). Other authors declare no competing interests.
Figures
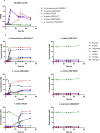


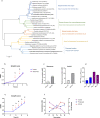
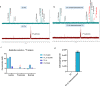

Similar articles
-
Phytate metabolism is mediated by microbial cross-feeding in the gut microbiota.Nat Microbiol. 2024 Jul;9(7):1812-1827. doi: 10.1038/s41564-024-01698-7. Epub 2024 Jun 10. Nat Microbiol. 2024. PMID: 38858593
-
Unravelling lactate-acetate and sugar conversion into butyrate by intestinal Anaerobutyricum and Anaerostipes species by comparative proteogenomics.Environ Microbiol. 2020 Nov;22(11):4863-4875. doi: 10.1111/1462-2920.15269. Epub 2020 Oct 12. Environ Microbiol. 2020. PMID: 33001550 Free PMC article.
-
Mutual Metabolic Interactions in Co-cultures of the Intestinal Anaerostipes rhamnosivorans With an Acetogen, Methanogen, or Pectin-Degrader Affecting Butyrate Production.Front Microbiol. 2019 Nov 1;10:2449. doi: 10.3389/fmicb.2019.02449. eCollection 2019. Front Microbiol. 2019. PMID: 31736896 Free PMC article.
-
Formation of propionate and butyrate by the human colonic microbiota.Environ Microbiol. 2017 Jan;19(1):29-41. doi: 10.1111/1462-2920.13589. Epub 2016 Dec 8. Environ Microbiol. 2017. PMID: 27928878 Review.
-
Phytate in foods and significance for humans: food sources, intake, processing, bioavailability, protective role and analysis.Mol Nutr Food Res. 2009 Sep;53 Suppl 2:S330-75. doi: 10.1002/mnfr.200900099. Mol Nutr Food Res. 2009. PMID: 19774556 Review.
Cited by
-
Analysis of differences in intestinal flora associated with different BMI status in colorectal cancer patients.J Transl Med. 2024 Feb 9;22(1):142. doi: 10.1186/s12967-024-04903-7. J Transl Med. 2024. PMID: 38331839 Free PMC article.
-
A unique gut microbiota signature in pulmonary arterial hypertension: A pilot study.Pulm Circ. 2022 Feb 28;12(1):e12051. doi: 10.1002/pul2.12051. eCollection 2022 Jan. Pulm Circ. 2022. PMID: 35506110 Free PMC article.
-
Analysis of the effects of β-mannanase on immune function and intestinal flora in broilers fed the low energy diet based on 16S rRNA sequencing and metagenomic sequencing.Poult Sci. 2024 May;103(5):103581. doi: 10.1016/j.psj.2024.103581. Epub 2024 Feb 22. Poult Sci. 2024. PMID: 38460218 Free PMC article.
-
Analysis of gut microbiotal diversity in healthy young adults in Sunan County, Gansu Province, China.Front Cell Infect Microbiol. 2023 May 24;13:1007505. doi: 10.3389/fcimb.2023.1007505. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37293209 Free PMC article.
-
Alterations in gut microbiota and urine metabolomics in infants with yin-deficiency constitution aged 0-2 years.Heliyon. 2023 Mar 24;9(4):e14684. doi: 10.1016/j.heliyon.2023.e14684. eCollection 2023 Apr. Heliyon. 2023. PMID: 37064462 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

