Fusion Peptide of SARS-CoV-2 Spike Rearranges into a Wedge Inserted in Bilayered Micelles
- PMID: 34375093
- PMCID: PMC8370118
- DOI: 10.1021/jacs.1c05435
Fusion Peptide of SARS-CoV-2 Spike Rearranges into a Wedge Inserted in Bilayered Micelles
Abstract
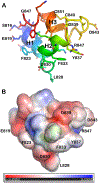
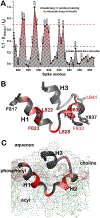
The receptor binding and proteolysis of Spike of SARS-CoV-2 release its S2 subunit to rearrange and catalyze viral-cell fusion. This deploys the fusion peptide for insertion into the cell membranes targeted. We show that this fusion peptide transforms from intrinsic disorder in solution into a wedge-shaped structure inserted in bilayered micelles, according to chemical shifts, 15N NMR relaxation, and NOEs. The globular fold of three helices contrasts the open, extended forms of this region observed in the electron density of compact prefusion states. In the hydrophobic, narrow end of the wedge, helices 1 and 2 contact the fatty acyl chains of phospholipids, according to NOEs and proximity to a nitroxide spin label deep in the membrane mimic. The polar end of the wedge may engage and displace lipid head groups and bind Ca2+ ions for membrane fusion. Polar helix 3 protrudes from the bilayer where it might be accessible to antibodies.
Figures

Similar articles
-
NMR structure and localization of a large fragment of the SARS-CoV fusion protein: Implications in viral cell fusion.Biochim Biophys Acta Biomembr. 2018 Feb;1860(2):407-415. doi: 10.1016/j.bbamem.2017.10.002. Epub 2017 Oct 5. Biochim Biophys Acta Biomembr. 2018. PMID: 28988778 Free PMC article.
-
Membranotropic and biological activities of the membrane fusion peptides from SARS-CoV spike glycoprotein: The importance of the complete internal fusion peptide domain.Biochim Biophys Acta Biomembr. 2021 Nov 1;1863(11):183697. doi: 10.1016/j.bbamem.2021.183697. Epub 2021 Jul 15. Biochim Biophys Acta Biomembr. 2021. PMID: 34274319 Free PMC article.
-
Interaction of Spike protein and lipid membrane of SARS-CoV-2 with Ursodeoxycholic acid, an in-silico analysis.Sci Rep. 2021 Nov 15;11(1):22288. doi: 10.1038/s41598-021-01705-5. Sci Rep. 2021. PMID: 34782703 Free PMC article.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
-
Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design.Viruses. 2021 Jan 14;13(1):109. doi: 10.3390/v13010109. Viruses. 2021. PMID: 33466921 Free PMC article. Review.
Cited by
-
Lassa virus glycoprotein complex review: insights into its unique fusion machinery.Biosci Rep. 2022 Feb 25;42(2):BSR20211930. doi: 10.1042/BSR20211930. Biosci Rep. 2022. PMID: 35088070 Free PMC article. Review.
-
Negatively charged residues in the membrane ordering activity of SARS-CoV-1 and -2 fusion peptides.Biophys J. 2022 Jan 18;121(2):207-227. doi: 10.1016/j.bpj.2021.12.024. Epub 2021 Dec 18. Biophys J. 2022. PMID: 34929193 Free PMC article.
-
Progressive membrane-binding mechanism of SARS-CoV-2 variant spike proteins.iScience. 2022 Aug 19;25(8):104722. doi: 10.1016/j.isci.2022.104722. Epub 2022 Jul 4. iScience. 2022. PMID: 35813872 Free PMC article.
-
Critical Negatively Charged Residues Are Important for the Activity of SARS-CoV-1 and SARS-CoV-2 Fusion Peptides.bioRxiv [Preprint]. 2021 Dec 6:2021.11.03.467161. doi: 10.1101/2021.11.03.467161. bioRxiv. 2021. Update in: Biophys J. 2022 Jan 18;121(2):207-227. doi: 10.1016/j.bpj.2021.12.024. PMID: 34909776 Free PMC article. Updated. Preprint.
-
A Suitable Membrane Distance Regulated by the RBD_ACE2 Interaction is Critical for SARS-CoV-2 Spike-Mediated Viral Invasion.Adv Sci (Weinh). 2023 Oct;10(28):e2301478. doi: 10.1002/advs.202301478. Epub 2023 Aug 17. Adv Sci (Weinh). 2023. PMID: 37590389 Free PMC article.
References
-
- Harrison SC Viral Membrane Fusion. Virology 2015, 479–480, 498–507. https://doi.org/10.1016/j.virol.2015.03.043. - DOI - PMC - PubMed
-
- Walls AC; Park Y-J; Tortorici MA; Wall A; McGuire AT; Veesler D Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181 (2), 281–292.e6. https://doi.org/10.1016/j.cell.2020.02.058. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

