Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area
- PMID: 34373850
- PMCID: PMC8325558
- DOI: 10.1016/j.wroa.2021.100111
Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area
Abstract
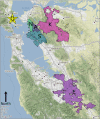
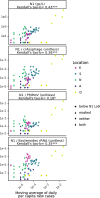
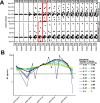
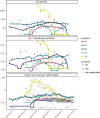
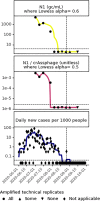
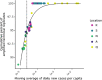
Wastewater surveillance for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA can be integrated with COVID-19 case data to inform timely pandemic response. However, more research is needed to apply and develop systematic methods to interpret the true SARS-CoV-2 signal from noise introduced in wastewater samples (e.g., from sewer conditions, sampling and extraction methods, etc.). In this study, raw wastewater was collected weekly from five sewersheds and one residential facility. The concentrations of SARS-CoV-2 in wastewater samples were compared to geocoded COVID-19 clinical testing data. SARS-CoV-2 was reliably detected (95% positivity) in frozen wastewater samples when reported daily new COVID-19 cases were 2.4 or more per 100,000 people. To adjust for variation in sample fecal content, four normalization biomarkers were evaluated: crAssphage, pepper mild mottle virus, Bacteroides ribosomal RNA (rRNA), and human 18S rRNA. Of these, crAssphage displayed the least spatial and temporal variability. Both unnormalized SARS-CoV-2 RNA signal and signal normalized to crAssphage had positive and significant correlation with clinical testing data (Kendall's Tau-b (τ)=0.43 and 0.38, respectively), but no normalization biomarker strengthened the correlation with clinical testing data. Locational dependencies and the date associated with testing data impacted the lead time of wastewater for clinical trends, and no lead time was observed when the sample collection date (versus the result date) was used for both wastewater and clinical testing data. This study supports that trends in wastewater surveillance data reflect trends in COVID-19 disease occurrence and presents tools that could be applied to make wastewater signal more interpretable and comparable across studies.
Keywords: Bacteroides; COVID-19; CrAssphage; Human 18S rRNA; Pepper mild mottle virus; Wastewater-based epidemiology.
© 2021 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Longitudinal and quantitative fecal shedding dynamics of SARS-CoV-2, pepper mild mottle virus, and crAssphage.mSphere. 2023 Aug 24;8(4):e0013223. doi: 10.1128/msphere.00132-23. Epub 2023 Jun 20. mSphere. 2023. PMID: 37338211 Free PMC article.
-
Assessment of seasonality and normalization techniques for wastewater-based surveillance in Ontario, Canada.Front Public Health. 2023 Aug 30;11:1186525. doi: 10.3389/fpubh.2023.1186525. eCollection 2023. Front Public Health. 2023. PMID: 37711234 Free PMC article.
-
Does normalization of SARS-CoV-2 concentrations by Pepper Mild Mottle Virus improve correlations and lead time between wastewater surveillance and clinical data in Alberta (Canada): comparing twelve SARS-CoV-2 normalization approaches.Sci Total Environ. 2023 Jan 15;856(Pt 1):158964. doi: 10.1016/j.scitotenv.2022.158964. Epub 2022 Sep 24. Sci Total Environ. 2023. PMID: 36167131 Free PMC article.
-
Assessment of crAssphage as a biological variable for SARS-CoV-2 data normalization in wastewater surveillance.J Appl Microbiol. 2024 Jul 2;135(7):lxae177. doi: 10.1093/jambio/lxae177. J Appl Microbiol. 2024. PMID: 39013607
-
Biomarkers selection for population normalization in SARS-CoV-2 wastewater-based epidemiology.Water Res. 2022 Sep 1;223:118985. doi: 10.1016/j.watres.2022.118985. Epub 2022 Aug 15. Water Res. 2022. PMID: 36030667 Free PMC article.
Cited by
-
Longitudinal fecal shedding of SARS-CoV-2, pepper mild mottle virus, and human mitochondrial DNA in COVID-19 patients.Front Med (Lausanne). 2024 Sep 11;11:1417967. doi: 10.3389/fmed.2024.1417967. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39323476 Free PMC article.
-
Advancing Public Health Surveillance: Integrating Modeling and GIS in the Wastewater-Based Epidemiology of Viruses, a Narrative Review.Pathogens. 2024 Aug 14;13(8):685. doi: 10.3390/pathogens13080685. Pathogens. 2024. PMID: 39204285 Free PMC article. Review.
-
COVID-19 trends at the University of Tennessee: predictive insights from raw sewage SARS-CoV-2 detection and evaluation and PMMoV as an indicator for human waste.Front Microbiol. 2024 Mar 28;15:1379194. doi: 10.3389/fmicb.2024.1379194. eCollection 2024. Front Microbiol. 2024. PMID: 38605711 Free PMC article.
-
Wastewater-based surveillance as a tool for public health action: SARS-CoV-2 and beyond.Clin Microbiol Rev. 2024 Mar 14;37(1):e0010322. doi: 10.1128/cmr.00103-22. Epub 2023 Dec 14. Clin Microbiol Rev. 2024. PMID: 38095438 Review.
-
Characterizing Spatial Information Loss for Wastewater Surveillance Using crAssphage: Effect of Decay, Temperature, and Population Mobility.Environ Sci Technol. 2023 Dec 12;57(49):20802-20812. doi: 10.1021/acs.est.3c05587. Epub 2023 Nov 28. Environ Sci Technol. 2023. PMID: 38015885
References
-
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. - DOI - PMC - PubMed
-
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. - DOI - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. - DOI - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., Simpson S.L., Thomas K.V., Verhagen R., Kitajima M., Mueller J.F., Korajkic A. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ. Res. 2020;110531 doi: 10.1016/j.envres.2020.110531. - DOI - PMC - PubMed
-
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

