HBx represses WDR77 to enhance HBV replication by DDB1-mediated WDR77 degradation in the liver
- PMID: 34373747
- PMCID: PMC8343998
- DOI: 10.7150/thno.57531
HBx represses WDR77 to enhance HBV replication by DDB1-mediated WDR77 degradation in the liver
Abstract
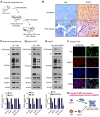
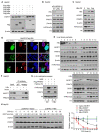
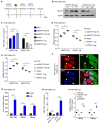
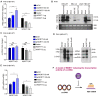
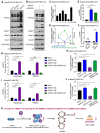
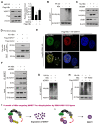
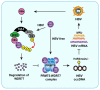
Rationale: Hepatitis B x protein (HBx) is required to initiate and maintain the replication of hepatitis B virus (HBV). Protein arginine methyltransferases 5 (PRMT5) negatively regulates HBV transcription. WD repeat domain 77 protein (WDR77) greatly enhances the methyltransferase activity of PRMT5. However, the role of WDR77 in the modulation of cccDNA transcription and HBV replication is poorly understood. In this study, we investigated the mechanism by which HBx modulated HBV replication involving WDR77 in the liver. Methods: A human liver-chimeric mouse model was established. Immunohistochemistry (IHC) staining, Western blot analysis, Southern blot analysis, Northern blot analysis, immunofluorescence assays, ELISA, RT-qPCR, CoIP assays, and ChIP assays were performed in human liver-chimeric mouse model, primary human hepatocytes (PHHs), HepG2-NTCP, dHepaRG and HepG2 cell lines. Results: HBV infection and HBx expression remarkably reduced the protein levels of WDR77 in human liver-chimeric mice and HepG2-NTCP cells. WDR77 restricted cccDNA transcription and HBV replication in PHHs and HepG2-NTCP cells. Mechanically, WDR77 enhanced PRMT5-triggered symmetric dimethylation of arginine 3 on H4 (H4R3me2s) on the cccDNA minichromosome to control cccDNA transcription. HBx drove the cellular DDB1-containing E3 ubiquitin ligase to degrade WDR77 through recruiting WDR77, leading to the disability of methyltransferase activity of PRMT5. Thus, HBx promoted HBV replication by driving a positive feedback loop of HBx-DDB1/WDR77/PRMT5/H4R3me2s/cccDNA/HBV/HBx in the liver. Conclusions: HBx attenuates the WDR77-mediated HBV repression by driving DDB1-induced WDR77 degradation in the liver. Our finding provides new insights into the mechanism by which HBx enhances HBV replication in the liver.
Keywords: DDB1; H4R3me2s; HBx; PRMT5; WDR77.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation.Hepatology. 2017 Aug;66(2):398-415. doi: 10.1002/hep.29133. Epub 2017 Jun 19. Hepatology. 2017. PMID: 28236308
-
Hepatitis B Virus HBx Protein Mediates the Degradation of Host Restriction Factors through the Cullin 4 DDB1 E3 Ubiquitin Ligase Complex.Cells. 2020 Mar 30;9(4):834. doi: 10.3390/cells9040834. Cells. 2020. PMID: 32235678 Free PMC article.
-
HAT1 signaling confers to assembly and epigenetic regulation of HBV cccDNA minichromosome.Theranostics. 2019 Sep 25;9(24):7345-7358. doi: 10.7150/thno.37173. eCollection 2019. Theranostics. 2019. PMID: 31695772 Free PMC article.
-
Identifying and Characterizing Interplay between Hepatitis B Virus X Protein and Smc5/6.Viruses. 2017 Apr 3;9(4):69. doi: 10.3390/v9040069. Viruses. 2017. PMID: 28368357 Free PMC article. Review.
-
Hepatitis B Viral Protein HBx: Roles in Viral Replication and Hepatocarcinogenesis.Viruses. 2024 Aug 26;16(9):1361. doi: 10.3390/v16091361. Viruses. 2024. PMID: 39339838 Free PMC article. Review.
Cited by
-
Pathogenicity and virulence of Hepatitis B virus.Virulence. 2022 Dec;13(1):258-296. doi: 10.1080/21505594.2022.2028483. Virulence. 2022. PMID: 35100095 Free PMC article. Review.
-
Elevated expression of WSB2 degrades p53 and activates the IGFBP3-AKT-mTOR-dependent pathway to drive hepatocellular carcinoma.Exp Mol Med. 2024 Feb;56(1):177-191. doi: 10.1038/s12276-023-01142-6. Epub 2024 Jan 4. Exp Mol Med. 2024. PMID: 38177295 Free PMC article.
-
Protein neddylation and its role in health and diseases.Signal Transduct Target Ther. 2024 Apr 5;9(1):85. doi: 10.1038/s41392-024-01800-9. Signal Transduct Target Ther. 2024. PMID: 38575611 Free PMC article. Review.
-
Hepatitis B x (HBx) as a Component of a Functional Cure for Chronic Hepatitis B.Biomedicines. 2022 Sep 7;10(9):2210. doi: 10.3390/biomedicines10092210. Biomedicines. 2022. PMID: 36140311 Free PMC article. Review.
-
Effects of neddylation on viral infection: an overview.Arch Virol. 2023 Dec 11;169(1):6. doi: 10.1007/s00705-023-05930-3. Arch Virol. 2023. PMID: 38081982 Review.
References
-
- Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486–500. - PubMed
-
- Revill P, Testoni B, Locarnini S, Zoulim F. Global strategies are required to cure and eliminate HBV infection. Nat Rev Gastroenterol Hepatol. 2016;13:239–48. - PubMed
-
- Benhenda S, Cougot D, Buendia MA, Neuveut C. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res. 2009;103:75–109. - PubMed
-
- Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

