Cytomegalovirus immediate-early 1 proteins form a structurally distinct protein class with adaptations determining cross-species barriers
- PMID: 34370791
- PMCID: PMC8376021
- DOI: 10.1371/journal.ppat.1009863
Cytomegalovirus immediate-early 1 proteins form a structurally distinct protein class with adaptations determining cross-species barriers
Abstract
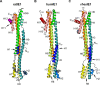
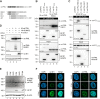
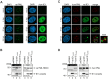
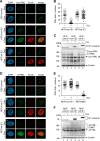
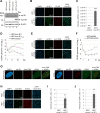
Restriction factors are potent antiviral proteins that constitute a first line of intracellular defense by blocking viral replication and spread. During co-evolution, however, viruses have developed antagonistic proteins to modulate or degrade the restriction factors of their host. To ensure the success of lytic replication, the herpesvirus human cytomegalovirus (HCMV) expresses the immediate-early protein IE1, which acts as an antagonist of antiviral, subnuclear structures termed PML nuclear bodies (PML-NBs). IE1 interacts directly with PML, the key protein of PML-NBs, through its core domain and disrupts the dot-like multiprotein complexes thereby abrogating the antiviral effects. Here we present the crystal structures of the human and rat cytomegalovirus core domain (IE1CORE). We found that IE1CORE domains, also including the previously characterized IE1CORE of rhesus CMV, form a distinct class of proteins that are characterized by a highly similar and unique tertiary fold and quaternary assembly. This contrasts to a marked amino acid sequence diversity suggesting that strong positive selection evolved a conserved fold, while immune selection pressure may have fostered sequence divergence of IE1. At the same time, we detected specific differences in the helix arrangements of primate versus rodent IE1CORE structures. Functional characterization revealed a conserved mechanism of PML-NB disruption, however, primate and rodent IE1 proteins were only effective in cells of the natural host species but not during cross-species infection. Remarkably, we observed that expression of HCMV IE1 allows rat cytomegalovirus replication in human cells. We conclude that cytomegaloviruses have evolved a distinct protein tertiary structure of IE1 to effectively bind and inactivate an important cellular restriction factor. Furthermore, our data show that the IE1 fold has been adapted to maximize the efficacy of PML targeting in a species-specific manner and support the concept that the PML-NBs-based intrinsic defense constitutes a barrier to cross-species transmission of HCMV.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

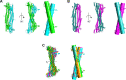
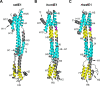
Similar articles
-
Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1-382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses.J Virol. 2015 Nov 11;90(3):1190-205. doi: 10.1128/JVI.01973-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26559840 Free PMC article.
-
The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation.J Virol. 2017 Jan 31;91(4):e02049-16. doi: 10.1128/JVI.02049-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27903803 Free PMC article.
-
Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions.PLoS Pathog. 2014 Nov 20;10(11):e1004512. doi: 10.1371/journal.ppat.1004512. eCollection 2014 Nov. PLoS Pathog. 2014. PMID: 25412268 Free PMC article.
-
The Human CMV IE1 Protein: An Offender of PML Nuclear Bodies.Adv Anat Embryol Cell Biol. 2017;223:77-94. doi: 10.1007/978-3-319-53168-7_4. Adv Anat Embryol Cell Biol. 2017. PMID: 28528440 Review.
-
Intrinsic cellular defense mechanisms targeting human cytomegalovirus.Virus Res. 2011 May;157(2):128-33. doi: 10.1016/j.virusres.2010.10.002. Epub 2010 Oct 8. Virus Res. 2011. PMID: 20934469 Review.
Cited by
-
Cross-Species Analysis of Innate Immune Antagonism by Cytomegalovirus IE1 Protein.Viruses. 2022 Jul 26;14(8):1626. doi: 10.3390/v14081626. Viruses. 2022. PMID: 35893691 Free PMC article.
-
Human Cytomegalovirus IE1 Impairs Neuronal Migration by Downregulating Connexin 43.J Virol. 2023 May 31;97(5):e0031323. doi: 10.1128/jvi.00313-23. Epub 2023 Apr 25. J Virol. 2023. PMID: 37097169 Free PMC article.
-
Daxx mediated histone H3.3 deposition on HSV-1 DNA restricts genome decompaction and the progression of immediate-early transcription.bioRxiv [Preprint]. 2024 Aug 15:2024.08.15.608064. doi: 10.1101/2024.08.15.608064. bioRxiv. 2024. PMID: 39185184 Free PMC article. Preprint.
-
IE1 of Human Cytomegalovirus Inhibits Necroptotic Cell Death via Direct and Indirect Modulation of the Necrosome Complex.Viruses. 2024 Feb 13;16(2):290. doi: 10.3390/v16020290. Viruses. 2024. PMID: 38400065 Free PMC article.
-
Triple lysine and nucleosome-binding motifs of the viral IE19 protein are required for human cytomegalovirus S-phase infections.mBio. 2024 Jun 12;15(6):e0016224. doi: 10.1128/mbio.00162-24. Epub 2024 May 2. mBio. 2024. PMID: 38695580 Free PMC article.
References
-
- Chemudupati M, Kenney AD, Bonifati S, Zani A, McMichael TM, Wu L, et al.. From APOBEC to ZAP: Diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochimica et biophysica acta Molecular cell research. 2019;1866(3):382–94. Epub 2018/10/06. doi: 10.1016/j.bbamcr.2018.09.012 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

