Subcellular Localization of Epstein-Barr Virus BLLF2 and Its Underlying Mechanisms
- PMID: 34367081
- PMCID: PMC8339435
- DOI: 10.3389/fmicb.2021.672192
Subcellular Localization of Epstein-Barr Virus BLLF2 and Its Underlying Mechanisms
Abstract
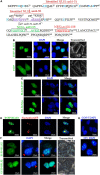
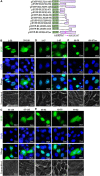
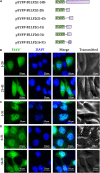
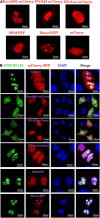
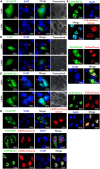
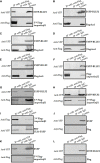
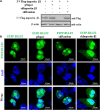
Epstein-Barr virus (EBV), the pathogen of several human malignancies, encodes many proteins required to be transported into the nucleus for viral DNA reproduction and nucleocapsids assembly in the lytic replication cycle. Here, fluorescence microscope, mutation analysis, interspecies heterokaryon assays, co-immunoprecipitation assay, RNA interference, and Western blot were performed to explore the nuclear import mechanism of EBV encoded BLLF2 protein. BLLF2 was shown to be a nucleocytoplasmic shuttling protein neither by a chromosomal region maintenance 1 (CRM1)- nor by a transporter associated with antigen processing (TAP)-dependent pathway. Yet, BLLF2's two functional nuclear localization signals (NLSs), NLS1 (16KRQALETVPHPQNRGR31) and NLS2 (44RRPRPPVAKRRRFPR58), were identified, whereas the predicted NES was nonfunctional. Finally, BLLF2 was proven to transport into the nucleus via a Ran-dependent and importin β1-dependent pathway. This mechanism may contribute to a more extensive insight into the assembly and synthesis of EBV virions in the nucleus, thus affording a new direction for the treatment of viruses.
Keywords: CRM1; EBV BLLF2; NES; NLS; TAP; importin.
Copyright © 2021 Li, Guo, Deng, Hu, Li, Deng, Zhong, Xie, Shi, Hong, Zheng, Cai and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Characterization of the Nucleocytoplasmic Transport Mechanisms of Epstein-Barr Virus BFLF2.Cell Physiol Biochem. 2018;51(4):1500-1517. doi: 10.1159/000495641. Epub 2018 Nov 29. Cell Physiol Biochem. 2018. PMID: 30497081
-
BGLF4 kinase modulates the structure and transport preference of the nuclear pore complex to facilitate nuclear import of Epstein-Barr virus lytic proteins.J Virol. 2015 Feb;89(3):1703-18. doi: 10.1128/JVI.02880-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410863 Free PMC article.
-
Characterization of nucleocytoplasmic shuttling and intracellular localization signals in Duck Enteritis Virus UL54.Biochimie. 2016 Aug;127:86-94. doi: 10.1016/j.biochi.2016.05.003. Epub 2016 May 5. Biochimie. 2016. PMID: 27157269
-
Nucleocytoplasmic protein transport and recycling of Ran.Cell Struct Funct. 1999 Dec;24(6):425-33. doi: 10.1247/csf.24.425. Cell Struct Funct. 1999. PMID: 10698256 Review.
-
Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review.
Cited by
-
Epstein-Barr Virus Envelope Glycoprotein gp110 Inhibits IKKi-Mediated Activation of NF-κB and Promotes the Degradation of β-Catenin.Microbiol Spectr. 2023 Jun 15;11(3):e0032623. doi: 10.1128/spectrum.00326-23. Epub 2023 Apr 6. Microbiol Spectr. 2023. PMID: 37022262 Free PMC article.
-
Multiple functions of the herpesvirus UL14 gene product in viral infection.Front Microbiol. 2024 Oct 23;15:1483022. doi: 10.3389/fmicb.2024.1483022. eCollection 2024. Front Microbiol. 2024. PMID: 39507342 Free PMC article. Review.
-
Modeling of horizontal pleiotropy identifies possible causal gene expression in systemic lupus erythematosus.Front Lupus. 2023;1:1234578. doi: 10.3389/flupu.2023.1234578. Epub 2023 Oct 3. Front Lupus. 2023. PMID: 37799268 Free PMC article.
References
-
- Belov G. A., Lidsky P. V., Mikitas O. V., Egger D., Lukyanov K. A., Bienz K., et al. (2004). Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 78 10166–10177. 10.1128/jvi.78.18.10166-10177.2004 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

