The Interactome of the VAP Family of Proteins: An Overview
- PMID: 34359948
- PMCID: PMC8306308
- DOI: 10.3390/cells10071780
The Interactome of the VAP Family of Proteins: An Overview
Abstract
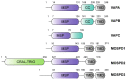
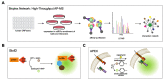
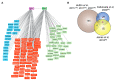
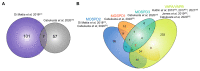
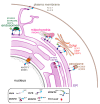
Membrane contact sites (MCS) are sites of close apposition of two organelles that help in lipid transport and synthesis, calcium homeostasis and several other biological processes. The VAMP-associated proteins (VAPs) VAPA, VAPB, MOSPD2 and the recently described MOSPD1 and MOSPD3 are tether proteins of MCSs that are mainly found at the endoplasmic reticulum (ER). VAPs interact with various proteins with a motif called FFAT (two phenylalanines in an acidic tract), recruiting the associated organelle to the ER. In addition to the conventional FFAT motif, the recently described FFNT (two phenylalanines in a neutral tract) and phospho-FFAT motifs contribute to the interaction with VAPs. In this review, we summarize and compare the recent interactome studies described for VAPs, including in silico and proximity labeling methods. Collectively, the interaction repertoire of VAPs is very diverse and highlights the complexity of interactions mediated by the different FFAT motifs to the VAPs.
Keywords: FFAT; MOSPD1; MOSPD2; MOSPD3; VAPA; VAPB; interactome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Human VAPome Analysis Reveals MOSPD1 and MOSPD3 as Membrane Contact Site Proteins Interacting with FFAT-Related FFNT Motifs.Cell Rep. 2020 Dec 8;33(10):108475. doi: 10.1016/j.celrep.2020.108475. Cell Rep. 2020. PMID: 33296653
-
What the VAP: The Expanded VAP Family of Proteins Interacting With FFAT and FFAT-Related Motifs for Interorganellar Contact.Contact (Thousand Oaks). 2021 May 9;4:25152564211012246. doi: 10.1177/25152564211012246. eCollection 2021 Jan 1. Contact (Thousand Oaks). 2021. PMID: 34036242 Free PMC article.
-
IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole.Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):12039-12044. doi: 10.1073/pnas.1709060114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078338 Free PMC article.
-
VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome.Biochim Biophys Acta. 2016 Aug;1861(8 Pt B):952-961. doi: 10.1016/j.bbalip.2016.02.009. Epub 2016 Feb 17. Biochim Biophys Acta. 2016. PMID: 26898182 Review.
-
VAP Proteins - From Organelle Tethers to Pathogenic Host Interactors and Their Role in Neuronal Disease.Front Cell Dev Biol. 2022 Jun 8;10:895856. doi: 10.3389/fcell.2022.895856. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35756994 Free PMC article. Review.
Cited by
-
Spinal cord neurone loss and foot placement changes in a rat knock-in model of amyotrophic lateral sclerosis Type 8.Brain Commun. 2024 May 24;6(3):fcae184. doi: 10.1093/braincomms/fcae184. eCollection 2024. Brain Commun. 2024. PMID: 38846532 Free PMC article.
-
Shared and Distinctive Neighborhoods of Emerin and Lamin B Receptor Revealed by Proximity Labeling and Quantitative Proteomics.J Proteome Res. 2022 Sep 2;21(9):2197-2210. doi: 10.1021/acs.jproteome.2c00281. Epub 2022 Aug 16. J Proteome Res. 2022. PMID: 35972904 Free PMC article.
-
ER as master regulator of membrane trafficking and organelle function.J Cell Biol. 2022 Oct 3;221(10):e202205135. doi: 10.1083/jcb.202205135. Epub 2022 Sep 15. J Cell Biol. 2022. PMID: 36108241 Free PMC article. Review.
-
Host MOSPD2 enrichment at the parasitophorous vacuole membrane varies between Toxoplasma strains and involves complex interactions.mSphere. 2023 Aug 24;8(4):e0067022. doi: 10.1128/msphere.00670-22. Epub 2023 Jun 21. mSphere. 2023. PMID: 37341482 Free PMC article.
-
In situ artificial contact sites (ISACS) between synthetic and endogenous organelle membranes allow for quantification of protein-tethering activities.J Biol Chem. 2022 May;298(5):101780. doi: 10.1016/j.jbc.2022.101780. Epub 2022 Feb 26. J Biol Chem. 2022. PMID: 35231443 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

